SARS-CoV-2-specific ELISA development
- PMID: 32780998
- PMCID: PMC7414735
- DOI: 10.1016/j.jim.2020.112832
SARS-CoV-2-specific ELISA development
Abstract
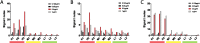
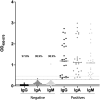
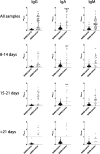
Critical to managing the spread of COVID-19 is the ability to diagnose infection and define the acquired immune response across the population. While genomic tests for the novel Several Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) detect the presence of viral RNA for a limited time frame, when the virus is shed in the upper respiratory tract, tests able to define exposure and infection beyond this short window of detectable viral replication are urgently needed. Following infection, antibodies are generated within days, providing a durable read-out and archive of exposure and infection. Several antibody tests have emerged to diagnose SARS-CoV-2. Here we report on a qualified quantitative ELISA assay that displays all the necessary characteristics for high-throughput sample analysis. Collectively, this test offers a quantitative opportunity to define both exposure and levels of immunity to SARS-CoV-2.
Copyright © 2020. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest Galit Alter is a founder of SeromYx.
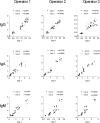
Figures




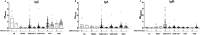



References
-
- Adams E.R. Evaluation of antibody testing for SARS-CoV-2 using ELISA and lateral flow immunoassay. medRxiv. 2020 doi: 10.1101/2020.04.15.20066407. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

