Aberrant BUB1 Overexpression Promotes Mitotic Segregation Errors and Chromosomal Instability in Multiple Myeloma
- PMID: 32781708
- PMCID: PMC7464435
- DOI: 10.3390/cancers12082206
Aberrant BUB1 Overexpression Promotes Mitotic Segregation Errors and Chromosomal Instability in Multiple Myeloma
Abstract
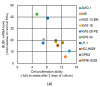
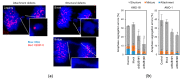
Chromosome instability (CIN), the hallmarks of cancer, reflects ongoing chromosomal changes caused by chromosome segregation errors and results in whole chromosomal or segmental aneuploidy. In multiple myeloma (MM), CIN contributes to the acquisition of tumor heterogeneity, and thereby, to disease progression, drug resistance, and eventual treatment failure; however, the underlying mechanism of CIN in MM remains unclear. Faithful chromosomal segregation is tightly regulated by a series of mitotic checkpoint proteins, such as budding uninhibited by benzimidazoles 1 (BUB1). In this study, we found that BUB1 was overexpressed in patient-derived myeloma cells, and BUB1 expression was significantly higher in patients in an advanced stage compared to those in an early stage. This suggested the involvement of aberrant BUB1 overexpression in disease progression. In human myeloma-derived cell lines (HMCLs), BUB1 knockdown reduced the frequency of chromosome segregation errors in mitotic cells. In line with this, partial knockdown of BUB1 showed reduced variations in chromosome number compared to parent cells in HMCLs. Finally, BUB1 overexpression was found to promote the clonogenic potency of HMCLs. Collectively, these results suggested that enhanced BUB1 expression caused an increase in mitotic segregation errors and the resultant emergence of subclones with altered chromosome numbers and, thus, was involved in CIN in MM.
Keywords: BUB1; chromosomal instability; chromosome segregation error; clonogenicity; multiple myeloma.
Conflict of interest statement
Y.F. is an employee of Nippon Shinyaku. J.K. has received research funding from Celgene, Kyowa Kirin, Chugai Pharmaceutical, Ono Pharmaceutical, Sanofi, Eisai, Bristol-Myers Squibb, Sysmex, Astellas Pharma, Pfizer, Dainippon Sumitomo Pharma, Nippon Shinyaku, Takeda, Shionogi, Asahi Kasei, Daiichi Sankyo, MSD, Taiho Pharmaceutical, Fujimoto Pharmaceutical, and Otsuka Pharmaceutical; honoraria from Janssen Pharmaceutical K.K, Celgene Corporation, Kyowa Kirin, Chugai Pharmaceutical, Ono Pharmaceutical, Takeda, Sanofi, Eisai, Bristol-Myers Squibb, Astellas Pharma, Pfizer, Nippon Shinyaku, Dainippon Sumitomo Pharma, Daiichi Sankyo, Fujimoto Pharmaceutical, Abbvie, and Otsuka Pharmaceutical; and is a consultant for Janssen Pharmaceutical K.K, Celgene, Bristol-Myers Squibb, Sanofi, and Abbvie. H.H. has received research funding from Celgene, Takeda, Ono Pharmaceutical, Kyowa Kirin, Sanofi, MSD, Chugai Pharmaceutical, Shionogi, Bayer, Eizai, and Astellas Pharma; honoraria from Janssen, Celgene, Takeda, Sanofi, and Ono; and is a consultant for Janssen, Takeda, and Celgene. M.T. has received research funding from Kyowa Kirin, Chugai Pharmaceutical, Eisai, and Astellas Pharma. T.K. has received honoraria from Chugai Pharmaceutical, Ono Pharmaceutical, Eisai, and Nippon Shinyaku. T.T. has received research funding from Nippon Shinyaku. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
-
- Walker B.A., Wardell C.P., Melchor L., Hulkki S., Potter N.E., Johnson D.C., Fenwick K., Kozarewa I., Gonzalez D., Lord C.J., et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood. 2012;120:1077–1086. doi: 10.1182/blood-2012-03-412981. - DOI - PubMed
-
- Walker B.A., Wardell C.P., Melchor L., Brioli A., Johnson D.C., Kaiser M.F., Mirabella F., Lopez-Corral L., Humphray S., Murray L., et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2014;28:384–390. doi: 10.1038/leu.2013.199. - DOI - PMC - PubMed
-
- Egan J.B., Shi C.X., Tembe W., Christoforides A., Kurdoglu A., Sinari S., Middha S., Asmann Y., Schmidt J., Braggio E., et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood. 2012;120:1060–1066. doi: 10.1182/blood-2012-01-405977. - DOI - PMC - PubMed
Grants and funding
- 26-A-4, 29-A-3/National Cancer Center Research and Development Funds
- H26-kakushin-teki-gan-ippan-074/Ministry of Health, Labour and Welfare of Japan
- JP16ck0106077h003, JP17ck0106348h0001, JP18ck0106348h0002 and JP19ck0106348h0003/Japan Agency for Medical Research and Development
- MEXT KAKENHI 16K09856, MEXT KAKENHI 18K08367/Ministry of Education, Culture, Sports, Science and Technology of Japan
- N.A./Japanese Society for Myeloma Research Award
LinkOut - more resources
Full Text Sources
Medical

