Spatial patterns of CTCF sites define the anatomy of TADs and their boundaries
- PMID: 32782014
- PMCID: PMC7422557
- DOI: 10.1186/s13059-020-02108-x
Spatial patterns of CTCF sites define the anatomy of TADs and their boundaries
Abstract
Background: Topologically associating domains (TADs) are genomic regions of self-interaction. Additionally, it is known that TAD boundaries are enriched in CTCF binding sites. In turn, CTCF sites are known to be asymmetric, whereby the convergent configuration of a pair of CTCF sites leads to the formation of a chromatin loop in vivo. However, to date, it has been unclear how to reconcile TAD structure with CTCF-based chromatin loops.
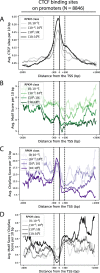
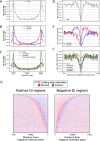
Results: We approach this problem by analysing CTCF binding site strengths and classifying clusters of CTCF sites along the genome on the basis of their relative orientation. Analysis of CTCF site orientation classes as a function of their spatial distribution along the human genome reveals that convergent CTCF site clusters are depleted while divergent CTCF clusters are enriched in the 5- to 100-kb range. We then analyse the distribution of CTCF binding sites as a function of TAD boundary conservation across seven primary human blood cell types. This reveals divergent CTCF site enrichment at TAD boundaries. Furthermore, convergent arrays of CTCF sites separate the left and right sections of TADs that harbour internal CTCF sites, resulting in unequal TAD 'halves'.
Conclusions: The orientation-based CTCF binding site cluster classification that we present reconciles TAD boundaries and CTCF site clusters in a mechanistically elegant fashion. This model suggests that the emergent structure of nuclear chromatin in the form of TADs relies on the obligate alternation of divergent and convergent CTCF site clusters that occur at different length scales along the genome.
Keywords: CTCF binding site clusters; CTCF orientation patterns; Chromatin architecture; Loop extrusion; TAD boundary conservation; TADs.
Conflict of interest statement
Not applicable.
Figures

Similar articles
-
Clustered CTCF binding is an evolutionary mechanism to maintain topologically associating domains.Genome Biol. 2020 Jan 7;21(1):5. doi: 10.1186/s13059-019-1894-x. Genome Biol. 2020. PMID: 31910870 Free PMC article.
-
Multi-feature clustering of CTCF binding creates robustness for loop extrusion blocking and Topologically Associating Domain boundaries.Nat Commun. 2023 Sep 12;14(1):5615. doi: 10.1038/s41467-023-41265-y. Nat Commun. 2023. PMID: 37699887 Free PMC article.
-
Pushing the TAD boundary: Decoding insulator codes of clustered CTCF sites in 3D genomes.Bioessays. 2024 Oct;46(10):e2400121. doi: 10.1002/bies.202400121. Epub 2024 Aug 21. Bioessays. 2024. PMID: 39169755 Review.
-
Insulation between adjacent TADs is controlled by the width of their boundaries through distinct mechanisms.Proc Natl Acad Sci U S A. 2025 Mar 18;122(11):e2413112122. doi: 10.1073/pnas.2413112122. Epub 2025 Mar 10. Proc Natl Acad Sci U S A. 2025. PMID: 40063813
-
The connection between BRG1, CTCF and topoisomerases at TAD boundaries.Nucleus. 2017 Mar 4;8(2):150-155. doi: 10.1080/19491034.2016.1276145. Epub 2017 Jan 6. Nucleus. 2017. PMID: 28060558 Free PMC article. Review.
Cited by
-
Modelling the bioinformatics tertiary analysis research process.BMC Bioinformatics. 2021 Sep 30;22(Suppl 13):452. doi: 10.1186/s12859-021-04310-5. BMC Bioinformatics. 2021. PMID: 34592928 Free PMC article.
-
CHD7 binds to insulators during neuronal differentiation.bioRxiv [Preprint]. 2025 Mar 29:2025.03.28.646031. doi: 10.1101/2025.03.28.646031. bioRxiv. 2025. PMID: 40196636 Free PMC article. Preprint.
-
Regulation of Transcription Factor NF-κB in Its Natural Habitat: The Nucleus.Cells. 2021 Mar 29;10(4):753. doi: 10.3390/cells10040753. Cells. 2021. PMID: 33805563 Free PMC article. Review.
-
CTCF-binding element regulates ESC differentiation via orchestrating long-range chromatin interaction between enhancers and HoxA.J Biol Chem. 2021 Jan-Jun;296:100413. doi: 10.1016/j.jbc.2021.100413. Epub 2021 Feb 11. J Biol Chem. 2021. PMID: 33581110 Free PMC article.
-
CTCF-mediated insulation and chromatin environment modulate Car5b escape from X inactivation.BMC Biol. 2025 Mar 3;23(1):68. doi: 10.1186/s12915-025-02137-7. BMC Biol. 2025. PMID: 40025499 Free PMC article.
References
-
- Atlasi Y, Megchelenbrink W, Peng T, Habibi E, Joshi O, Wang SY, et al. Epigenetic modulation of a hardwired 3D chromatin landscape in two naive states of pluripotency. Nat Cell Biol. 2019;21:568–578. - PubMed
-
- Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

