Constitutive immune mechanisms: mediators of host defence and immune regulation
- PMID: 32782357
- PMCID: PMC7418297
- DOI: 10.1038/s41577-020-0391-5
Constitutive immune mechanisms: mediators of host defence and immune regulation
Abstract
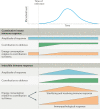
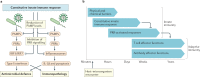
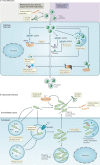
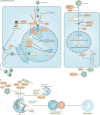
The immune system enables organisms to combat infections and to eliminate endogenous challenges. Immune responses can be evoked through diverse inducible pathways. However, various constitutive mechanisms are also required for immunocompetence. The inducible responses of pattern recognition receptors of the innate immune system and antigen-specific receptors of the adaptive immune system are highly effective, but they also have the potential to cause extensive immunopathology and tissue damage, as seen in many infectious and autoinflammatory diseases. By contrast, constitutive innate immune mechanisms, including restriction factors, basal autophagy and proteasomal degradation, tend to limit immune responses, with loss-of-function mutations in these pathways leading to inflammation. Although they function through a broad and heterogeneous set of mechanisms, the constitutive immune responses all function as early barriers to infection and aim to minimize any disruption of homeostasis. Supported by recent human and mouse data, in this Review we compare and contrast the inducible and constitutive mechanisms of immunosurveillance.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. - PubMed
-
- van der Poll T, van de Veerdonk FL, Scicluna BP, Netea MG. The immunopathology of sepsis and potential therapeutic targets. Nat. Rev. Immunol. 2017;17:407–420. - PubMed
-
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

