An optimal prognostic model based on gene expression for clear cell renal cell carcinoma
- PMID: 32782559
- PMCID: PMC7400162
- DOI: 10.3892/ol.2020.11780
An optimal prognostic model based on gene expression for clear cell renal cell carcinoma
Abstract
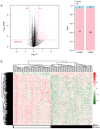
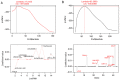
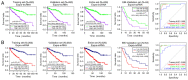
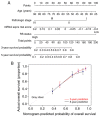
Clear cell renal cell carcinoma (ccRCC) is the most prevalent type of RCC; however, prognostic prediction tools for ccRCC are scant. Developing mRNA or long non-coding RNA (lncRNA)-based risk assessment tools may improve the prognosis in patients with ccRCC. RNA-sequencing and prognostic data from patients with ccRCC were downloaded from The Cancer Genome Atlas and the European Bioinformatics Institute Array database at the National Center for Biotechnology Information. Differentially expressed (DE) RNAs (DERs) and prognostic DERs were screened between less favorable and favorable prognoses using the limma package in R 3.4.1, and analyzed using univariate and multivariate Cox regression analyses, respectively. Risk score models were constructed using optimal combinations of DEmRNAs and DElncRNAs identified using the Least Absolute Shrinkage And Selection Operator Cox regression model of the penalized package. Associations between risk score models and overall survival time were evaluated. Independent prognostic clinical factors were screened using univariate and multivariate Cox regression analyses, and nomogram models were constructed. Gene Ontology biological processes and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were conducted using the clusterProfiler package in R3.4.1. A total of 451 DERs were identified, including 404 mRNAs and 47 lncRNAs, between less favorable and favorable prognoses, and 269 DERs, including 233 mRNAs and 36 lncRNAs, were identified as independent prognostic factors. Optimal combinations including 10 DEmRNAs or 10 DElncRNAs were screened using four risk score models based on the status or expression levels of the 10 DEmRNAs or 10 DElncRNAs. The model based on the expression levels of the 10 DEmRNAs had the highest prognostic power. These prognostic DEmRNAs may be involved in biological processes associated with the inflammatory response, complement and coagulation cascades and neuroactive ligand-receptor interaction pathways. The present validated risk assessment tool based on the expression levels of these 10 DEmRNAs may help to identify patients with ccRCC at a high risk of mortality. These 10 DEmRNAs in optimal combinations may serve as prognostic biomarkers and help to elucidate the pathogenesis of ccRCC.
Keywords: DEGs; lncRNAs; pathway enrichment analysis; prognostic model; risk score.
Copyright: © Xu et al.
Figures



References
LinkOut - more resources
Full Text Sources
