The SNARE regulator Complexin3 is a target of the cone circadian clock
- PMID: 32783205
- PMCID: PMC8190822
- DOI: 10.1002/cne.25004
The SNARE regulator Complexin3 is a target of the cone circadian clock
Abstract
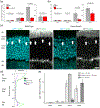
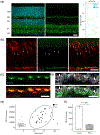
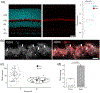
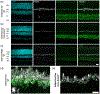
BMAL1 is a core component of the mammalian circadian clockwork. Removal of BMAL1 from the retina significantly affects visual information processing in both rod and cone pathways. To identify potential pathways and/or molecules through which BMAL1 alters signal transmission at the cone pedicle, we performed an RNA-seq differential expression analysis between cone-specific Bmal1 knockout cones (cone-Bmal1-/- ) and wild-type (WT) cones. We found 88 genes differentially expressed. Among these, Complexin3 (Cplx3), a SNARE regulator at ribbon synapses, was downregulated fivefold in the mutant cones. The purpose of this work was to determine whether BMAL1 and/or the cone clock controls CPLX3 protein expression at cone pedicles. We found that CPLX3 expression level was decreased twofold in cone-Bmal1-/- cones. Furthermore, CPLX3 expression was downregulated at night compared to the day in WT cones but remained constitutively low in mutant cones both day and night. The transcript and protein expression levels of Cplx4-the other complexin expressed in cones-were similar in WT and mutant cones; in WT cones, CPLX4 protein level did not change with the time of day. In silico analysis revealed four potential BMAL1:CLOCK binding sites upstream from exon one of Cplx3 and none upstream of exon one of Cplx4. Our results suggest that CPLX3 expression is regulated at the transcriptional level by the cone clock. The modulation of CPLX3 may be a mechanism by which the clock controls the cone synaptic transfer function to second-order cells and thereby impacts retinal signal processing during the day/night cycle.
Keywords: Bmal1; Cplx3; Cplx4; SNARE proteins; circadian clock; cones; retina; ribbon synapses.
© 2020 Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Figures



References
-
- Baba K, Piano I, Lyuboslavsky P, Chrenek MA, Sellers JT, Zhang S, … Iuvone PM (2018). Removal of clock gene Bmal1 from the retina affects retinal development and accelerates cone photoreceptor degeneration during aging. Proceedings of the National Academy of Sciences of the United States of America, 115(51), 13099–13104. 10.1073/pnas.1808137115 - DOI - PMC - PubMed
-
- Baba K, Ribelayga CP, Michael Iuvone P, & Tosini G (2018). The retinal circadian clock and photoreceptor viability. In Ash JD, Anderson RE, LaVail MM, Bowes Rickman C, Hollyfield JG, & Grimm C (Eds.), Retinal degenerative diseases (Vol. 1074, pp. 345–350). New York: Springer International Publishing. 10.1007/978-3-319-75402-4_42 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

