Joint testing of donor and recipient genetic matching scores and recipient genotype has robust power for finding genes associated with transplant outcomes
- PMID: 32783273
- PMCID: PMC7658035
- DOI: 10.1002/gepi.22349
Joint testing of donor and recipient genetic matching scores and recipient genotype has robust power for finding genes associated with transplant outcomes
Abstract
Genetic matching between transplant donor and recipient pairs has traditionally focused on the human leukocyte antigen (HLA) regions of the genome, but recent studies suggest that matching for non-HLA regions may be important as well. We assess four genetic matching scores for use in association analyses of transplant outcomes. These scores describe genetic ancestry distance using identity-by-state, or genetic incompatibility or mismatch of the two genomes and therefore may reflect different underlying biological mechanisms for donor and recipient genes to influence transplant outcomes. Our simulation studies show that jointly testing these scores with the recipient genotype is a powerful method for preliminary screening and discovery of transplant outcome related single nucleotide polymorphisms (SNPs) and gene regions. Following these joint tests with marginal testing of the recipient genotype and matching score separately can lead to further understanding of the biological mechanisms behind transplant outcomes. In addition, we present results of a liver transplant data analysis that shows joint testing can detect SNPs significantly associated with acute rejection in liver transplant.
Keywords: genetic matching scores; joint testing; transplant genetics.
© 2020 Wiley Periodicals LLC.
Conflict of interest statement
Conflict of Interests
The authors declare that there are no conflicts of interest.
Figures

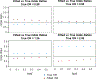
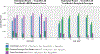


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

