New anti-viral drugs for the treatment of COVID-19 instead of favipiravir
- PMID: 32783586
- PMCID: PMC7484583
- DOI: 10.1080/07391102.2020.1806112
New anti-viral drugs for the treatment of COVID-19 instead of favipiravir
Abstract
The SARS-CoV-2 virus is a major problem in the world right now. Currently, all the attention of research centers and governments globally are focused on the investigation of vaccination studies and the discovery of small molecules that inhibit the SARS-CoV-2 virus in the treatment of patients. The goal of this study was to locate small molecules to be used against COVID19 instead of favipiravir. Favipiravir analogues were selected as drug candidates from the PubChem web tool. The RNA dependent RNA polymerase (RdRp) protein was selected as the target protein as favipiravir inhibits this protein in the human body. Initially, the inhibition activity of the studied compounds against RdRp of different virus types was investigated. Then, the inhibition properties of selected drug candidates and favipiravir were examined in detail against SARS-CoV-2 RdRp proteins. It was found that 2-oxo-1H-pyrazine-3-carboxamide performed better than favipiravir in the results of molecular docking, molecular mechanics Poisson-Boltzmann surface area (MM-PSBA) calculations, and ADME analyses.Communicated by Ramaswamy H. Sarma.
Keywords: ADME; COVID19; MM-PBSA; RNA polymerase; favipiravir.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures





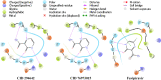

References
-
- Beylkin, D., Kumar, G., Zhou, W., Park, J., Jeevan, T., Lagisetti, C., Harfoot, R., Webby, R. J., White, S. W., & Webb, T. R. (2017). Protein-structure assisted optimization of 4, 5-dihydroxypyrimidine-6-carboxamide inhibitors of influenza virus endonuclease. Scientific Reports, 7(1), 1–12. 10.1038/s41598-017-17419-6 - DOI - PMC - PubMed
-
- Bıçak, B., Gündüz, S. K., Kökcü, Y., Özel, A. E., & Akyüz, S. (2019). Molecular docking and molecular dynamics studies of L-glycyl-L-glutamic acid dipeptide. Bilge International Journal of Science and Technology Research, 3(1), 1–9. 10.30516/bilgesci.476841 - DOI
-
- Bost, A. G., Carnahan, R. H., Lu, X. T., & Denison, M. R. (2000). Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. Journal of Virology, 74(7), 3379–3387. 10.1021/acs.biochem.5b01087 10.1128/JVI.74.7.3379-3387.2000 - DOI - DOI - PMC - PubMed
-
- Brockway, S. M., Clay, C. T., Lu, X. T., & Denison, M. R. (2003). Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. Journal of Virology, 77(19), 10515–10527. 10.1128/jvi.77.19.10515-10527.2003 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
