Single-Cell Sequencing of Peripheral Mononuclear Cells Reveals Distinct Immune Response Landscapes of COVID-19 and Influenza Patients
- PMID: 32783921
- PMCID: PMC7368915
- DOI: 10.1016/j.immuni.2020.07.009
Single-Cell Sequencing of Peripheral Mononuclear Cells Reveals Distinct Immune Response Landscapes of COVID-19 and Influenza Patients
Abstract
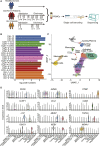
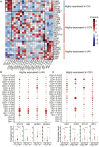
The coronavirus disease 2019 (COVID-19) pandemic poses a current world-wide public health threat. However, little is known about its hallmarks compared to other infectious diseases. Here, we report the single-cell transcriptional landscape of longitudinally collected peripheral blood mononuclear cells (PBMCs) in both COVID-19- and influenza A virus (IAV)-infected patients. We observed increase of plasma cells in both COVID-19 and IAV patients and XIAP associated factor 1 (XAF1)-, tumor necrosis factor (TNF)-, and FAS-induced T cell apoptosis in COVID-19 patients. Further analyses revealed distinct signaling pathways activated in COVID-19 (STAT1 and IRF3) versus IAV (STAT3 and NFκB) patients and substantial differences in the expression of key factors. These factors include relatively increase of interleukin (IL)6R and IL6ST expression in COVID-19 patients but similarly increased IL-6 concentrations compared to IAV patients, supporting the clinical observations of increased proinflammatory cytokines in COVID-19 patients. Thus, we provide the landscape of PBMCs and unveil distinct immune response pathways in COVID-19 and IAV patients.
Keywords: COVID-19; IL-6R; apoptosis; influenza; scRNA-seq.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests Employees of BGI have stock holdings in BGI.
Figures


Comment in
-
The interplay between dendritic cells and CD8 T lymphocytes is a crucial component of SARS-CoV-2 immunity.Cell Mol Immunol. 2021 Feb;18(2):247-249. doi: 10.1038/s41423-020-00624-1. Epub 2021 Jan 8. Cell Mol Immunol. 2021. PMID: 33420356 Free PMC article. No abstract available.
-
COVID-19 and flu: conserved or specific immune signature?Cell Mol Immunol. 2021 Feb;18(2):245-246. doi: 10.1038/s41423-020-00595-3. Epub 2021 Jan 11. Cell Mol Immunol. 2021. PMID: 33432063 Free PMC article. No abstract available.
References
-
- Desai P., Tahiliani V., Hutchinson T.E., Dastmalchi F., Stanfield J., Abboud G., Thomas P.G., Ware C.F., Song J., Croft M., Salek-Ardakani S. The TNF superfamily molecule LIGHT promotes the generation of circulating and lung-resident memory CD8 T cells following an acute respiratory virus infection. J. Immunol. 2018;200:2894–2904. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

