Aedes aegypti from Amazon Basin Harbor High Diversity of Novel Viral Species
- PMID: 32784421
- PMCID: PMC7472207
- DOI: 10.3390/v12080866
Aedes aegypti from Amazon Basin Harbor High Diversity of Novel Viral Species
Abstract
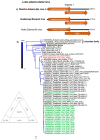
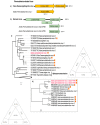
Viruses are the most diverse and abundant microorganisms on earth, highly adaptive to a wide range of hosts. Viral diversity within invertebrate hosts has gained notoriety in recent years in public health as several such viruses have been of medical importance. Aedes aegypti serves as a vector for several viruses that have caused epidemics within the last year throughout Brazil; including Dengue, Zika and Chikungunya. This study aimed to identify new viral agents within Aedes aegypti mosquito in a city of the Amazonian region, where it is highly endemic. Metagenomic investigation was performed on 60 mosquito pools and viral RNA sequences present in their microbiota were characterized using genomic and phylogenetic tools. In total, we identified five putative novel virus species related to the Sobemovirus genus, Iflavirus genus and Permutatetraviridae family. These findings indicate a diverse taxonomy of viruses present in the mosquito microbiota of the Amazon, the region with the greatest invertebrate diversity in the world.
Keywords: Aedes aegypti; Amazon; NGS; insect-virus; microbiota; mosquito; viral diversity.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

