MicroRNA-222 Regulates Melanoma Plasticity
- PMID: 32784455
- PMCID: PMC7464186
- DOI: 10.3390/jcm9082573
MicroRNA-222 Regulates Melanoma Plasticity
Abstract
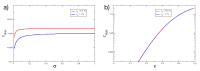
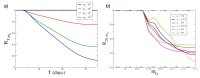
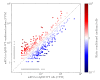
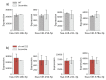
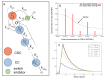
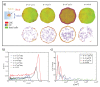
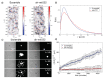
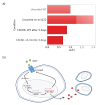
Melanoma is one of the most aggressive and highly resistant tumors. Cell plasticity in melanoma is one of the main culprits behind its metastatic capabilities. The detailed molecular mechanisms controlling melanoma plasticity are still not completely understood. Here we combine mathematical models of phenotypic switching with experiments on IgR39 human melanoma cells to identify possible key targets to impair phenotypic switching. Our mathematical model shows that a cancer stem cell subpopulation within the tumor prevents phenotypic switching of the other cancer cells. Experiments reveal that hsa-mir-222 is a key factor enabling this process. Our results shed new light on melanoma plasticity, providing a potential target and guidance for therapeutic studies.
Keywords: cancer stem cells; hsa-mir-222; kinetic equations; melanoma; phenotypic switching; reaction diffusion; tumor plasticity.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Tetteh P.W., Basak O., Farin H.F., Wiebrands K., Kretzschmar K., Begthel H., van den Born M., Korving J., de Sauvage F., van Es J.H., et al. Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. - DOI - PubMed

