SARS-CoV-2 Entry Inhibitors: Small Molecules and Peptides Targeting Virus or Host Cells
- PMID: 32784899
- PMCID: PMC7460888
- DOI: 10.3390/ijms21165707
SARS-CoV-2 Entry Inhibitors: Small Molecules and Peptides Targeting Virus or Host Cells
Abstract
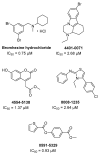
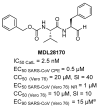
The pandemic evolution of SARS-CoV-2 infection is forcing the scientific community to unprecedented efforts to explore all possible approaches against COVID-19. In this context, targeting virus entry is a promising antiviral strategy for controlling viral infections. The main strategies pursued to inhibit the viral entry are considering both the virus and the host factors involved in the process. Primarily, direct-acting antivirals rely on inhibition of the interaction between ACE2 and the receptor binding domain (RBD) of the Spike (S) protein or targeting the more conserved heptad repeats (HRs), involved in the membrane fusion process. The inhibition of host TMPRSS2 and cathepsins B/L may represent a complementary strategy to be investigated. In this review, we discuss the development entry inhibitors targeting the S protein, as well as the most promising host targeting strategies involving TMPRSS2 and CatB/L, which have been exploited so far against CoVs and other related viruses.
Keywords: COVID-19; SARS-CoV-2 entry inhibitors; TMPRSS2; cathepsins; coronavirus; peptides inhibitors; small molecules inhibitors; spike.
Conflict of interest statement
The authors declare no conflict of interest
Figures


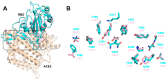





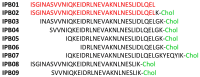







References
-
- WHO . Ten Threats to Global Health in 2019. WHO; Geneva, Switzerland: 2019. [(accessed on 8 June 2020)]. Available online: https://www.who.int/emergencies/ten-threats-to-global-health-in-2019.
-
- WHO . Prioritizing Diseases for Research and Development in Emergency Contexts. WHO; Geneva, Switzerland: 2020. [(accessed on 8 June 2020)]. Available online: Https://www.who.int/activities/prioritizing-diseases-for-research-anddev....
-
- Rolling Updates on Coronavirus Disease (COVID-19) [(accessed on 8 June 2020)]; Available online: Https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-a....
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

