The Evolution of Duplicated Genes of the Cpi-17/Phi-1 (ppp1r14) Family of Protein Phosphatase 1 Inhibitors in Teleosts
- PMID: 32784920
- PMCID: PMC7460850
- DOI: 10.3390/ijms21165709
The Evolution of Duplicated Genes of the Cpi-17/Phi-1 (ppp1r14) Family of Protein Phosphatase 1 Inhibitors in Teleosts
Abstract
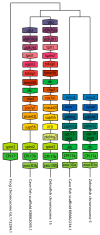
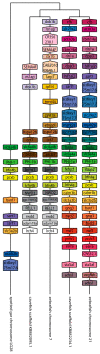
The Cpi-17 (ppp1r14) gene family is an evolutionarily conserved, vertebrate specific group of protein phosphatase 1 (PP1) inhibitors. When phosphorylated, Cpi-17 is a potent inhibitor of myosin phosphatase (MP), a holoenzyme complex of the regulatory subunit Mypt1 and the catalytic subunit PP1. Myosin phosphatase dephosphorylates the regulatory myosin light chain (Mlc2) and promotes actomyosin relaxation, which in turn, regulates numerous cellular processes including smooth muscle contraction, cytokinesis, cell motility, and tumor cell invasion. We analyzed zebrafish homologs of the Cpi-17 family, to better understand the mechanisms of myosin phosphatase regulation. We found single homologs of both Kepi (ppp1r14c) and Gbpi (ppp1r14d) in silico, but we detected no expression of these genes during early embryonic development. Cpi-17 (ppp1r14a) and Phi-1 (ppp1r14b) each had two duplicate paralogs, (ppp1r14aa and ppp1r14ab) and (ppp1r14ba and ppp1r14bb), which were each expressed during early development. The spatial expression pattern of these genes has diverged, with ppp1r14aa and ppp1r14bb expressed primarily in smooth muscle and skeletal muscle, respectively, while ppp1r14ab and ppp1r14ba are primarily expressed in neural tissue. We observed that, in in vitro and heterologous cellular systems, the Cpi-17 paralogs both acted as potent myosin phosphatase inhibitors, and were indistinguishable from one another. In contrast, the two Phi-1 paralogs displayed weak myosin phosphatase inhibitory activity in vitro, and did not alter myosin phosphorylation in cells. Through deletion and chimeric analysis, we identified that the difference in specificity for myosin phosphatase between Cpi-17 and Phi-1 was encoded by the highly conserved PHIN (phosphatase holoenzyme inhibitory) domain, and not the more divergent N- and C- termini. We also showed that either Cpi-17 paralog can rescue the knockdown phenotype, but neither Phi-1 paralog could do so. Thus, we provide new evidence about the biochemical and developmental distinctions of the zebrafish Cpi-17 protein family.
Keywords: Cpi-17 (ppp1r14a); Danio rerio; Mypt1; PP1; Phi-1 (ppp1r14b); genome duplication.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

