Centromeric Transcription: A Conserved Swiss-Army Knife
- PMID: 32784923
- PMCID: PMC7463856
- DOI: 10.3390/genes11080911
Centromeric Transcription: A Conserved Swiss-Army Knife
Abstract
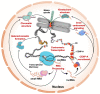
In most species, the centromere is comprised of repetitive DNA sequences, which rapidly evolve. Paradoxically, centromeres fulfill an essential function during mitosis, as they are the chromosomal sites wherein, through the kinetochore, the mitotic spindles bind. It is now generally accepted that centromeres are transcribed, and that such transcription is associated with a broad range of functions. More than a decade of work on this topic has shown that centromeric transcripts are found across the eukaryotic tree and associate with heterochromatin formation, chromatin structure, kinetochore structure, centromeric protein loading, and inner centromere signaling. In this review, we discuss the conservation of small and long non-coding centromeric RNAs, their associations with various centromeric functions, and their potential roles in disease.
Keywords: centromere; chromatin; epigenetics; non-coding RNAs; nucleosomes; transcription.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures


References
-
- Melters D.P., Bradnam K.R., Young H.A., Telis N., May M.R., Ruby J.G., Sebra R., Peluso P., Eid J., Rank D., et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013;14:R10. doi: 10.1186/gb-2013-14-1-r10. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

