The prevalence and genomic context of Shiga toxin 2a genes in E. coli found in cattle
- PMID: 32785271
- PMCID: PMC7423110
- DOI: 10.1371/journal.pone.0232305
The prevalence and genomic context of Shiga toxin 2a genes in E. coli found in cattle
Abstract
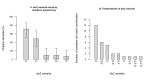
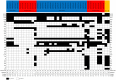
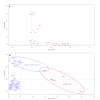
Shiga toxin-producing Escherichia coli (STEC) that cause severe disease predominantly carry the toxin gene variant stx2a. However, the role of Shiga toxin in the ruminant reservoirs of this zoonotic pathogen is poorly understood and strains that cause severe disease in humans (HUSEC) likely constitute a small and atypical subset of the overall STEC flora. The aim of this study was to investigate the presence of stx2a in samples from cattle and to isolate and characterize stx2a-positive E. coli. In nationwide surveys in Sweden and Norway samples were collected from individual cattle or from cattle herds, respectively. Samples were tested for Shiga toxin genes by real-time PCR and amplicon sequencing and stx2a-positive isolates were whole genome sequenced. Among faecal samples from Sweden, stx1 was detected in 37%, stx2 in 53% and stx2a in 5% and in skin (ear) samples in 64%, 79% and 2% respectively. In Norway, 79% of the herds were positive for stx1, 93% for stx2 and 17% for stx2a. Based on amplicon sequencing the most common stx2 types in samples from Swedish cattle were stx2a and stx2d. Multilocus sequence typing (MLST) of 39 stx2a-positive isolates collected from both countries revealed substantial diversity with 19 different sequence types. Only a few classical LEE-positive strains similar to HUSEC were found among the stx2a-positive isolates, notably a single O121:H19 and an O26:H11. Lineages known to include LEE-negative HUSEC were also recovered including, such as O113:H21 (sequence type ST-223), O130:H11 (ST-297), and O101:H33 (ST-330). We conclude that E. coli encoding stx2a in cattle are ranging from strains similar to HUSEC to unknown STEC variants. Comparison of isolates from human HUS cases to related STEC from the ruminant reservoirs can help identify combinations of virulence attributes necessary to cause HUS, as well as provide a better understanding of the routes of infection for rare and emerging pathogenic STEC.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Spinale JM, Ruebner RL, Copelovitch L, Kaplan BS. Long-term outcomes of Shiga toxin hemolytic uremic syndrome. Pediatr Nephrol Berl Ger. 2013. November;28(11):2097–105. - PubMed
-
- Koutsoumanis K, Allende A, Alvarez-Ordóñez A, Bover-Cid S, Chemaly M, Davies R, et al. Pathogenicity assessment of Shiga toxin-producing Escherichia coli (STEC) and the public health risk posed by contamination of food with STEC. EFSA J. 2020;18(1):e05967.
-
- Tozzoli R, Grande L, Michelacci V, Ranieri P, Maugliani A, Caprioli A, et al. Shiga toxin-converting phages and the emergence of new pathogenic Escherichia coli: a world in motion. Front Cell Infect Microbiol [Internet]. 2014. June 20 [cited 2014 Nov 4];4 Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4064290/ - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

