Macrophage morphology correlates with single-cell diversity and prognosis in colorectal liver metastasis
- PMID: 32785653
- PMCID: PMC7596819
- DOI: 10.1084/jem.20191847
Macrophage morphology correlates with single-cell diversity and prognosis in colorectal liver metastasis
Abstract
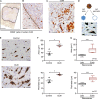
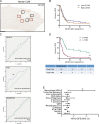
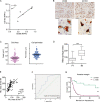
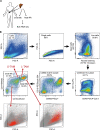
It has long been known that in vitro polarized macrophages differ in morphology. Stemming from a conventional immunohistology observation, we set out to test the hypothesis that morphology of tumor-associated macrophages (TAMs) in colorectal liver metastasis (CLM) represents a correlate of functional diversity with prognostic significance. Density and morphological metrics of TAMs were measured and correlated with clinicopathological variables. While density of TAMs did not correlate with survival of CLM patients, the cell area identified small (S-TAM) and large (L-TAM) macrophages that were associated with 5-yr disease-free survival rates of 27.8% and 0.2%, respectively (P < 0.0001). RNA sequencing of morphologically distinct macrophages identified LXR/RXR as the most enriched pathway in large macrophages, with up-regulation of genes involved in cholesterol metabolism, scavenger receptors, MERTK, and complement. In single-cell analysis of mononuclear phagocytes from CLM tissues, S-TAM and L-TAM signatures were differentially enriched in individual clusters. These results suggest that morphometric characterization can serve as a simple readout of TAM diversity with strong prognostic significance.
© 2020 Donadon et al.
Conflict of interest statement
Disclosures: A. Mantovani reported personal fees from Ventana, Pierre Fabre, Verily, AbbVie, Astra Zeneca, Verseau Therapeutics, Compugen, Myeloid Therapeutics, Third Rock Venture, Imcheck Therapeutics, Ellipses, Novartis, Roche, Macrophage Pharma, Biovelocita, Merck, and Principia; grants from Novartis; and "other" from Cedarlane Laboratories Ltd, Hycult Biotechnology, eBioscience, BioLegend, ABCAM Plc, Novus Biologicals, Enzo Life (ex-Alexis Corp.), and Affymetrix outside the submitted work. He is the inventor of patents related to PTX3 and other innate immunity molecules and receives royalties for reagents related to innate immunity. No other disclosures were reported.
Figures



Comment in
-
Cancer metastasis linked to macrophage size, shape, and metabolism.J Exp Med. 2020 Nov 2;217(11):e20201259. doi: 10.1084/jem.20201259. J Exp Med. 2020. PMID: 32997733 Free PMC article.
References
-
- A-Gonzalez N., Quintana J.A., García-Silva S., Mazariegos M., González de la Aleja A., Nicolás-Ávila J.A., Walter W., Adrover J.M., Crainiciuc G., Kuchroo V.K., et al. . 2017. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med. 214:1281–1296. 10.1084/jem.20161375 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

