Characterization of an Aggregated Three-Dimensional Cell Culture Model by Multimodal Mass Spectrometry Imaging
- PMID: 32786495
- PMCID: PMC7497704
- DOI: 10.1021/acs.analchem.0c02389
Characterization of an Aggregated Three-Dimensional Cell Culture Model by Multimodal Mass Spectrometry Imaging
Abstract
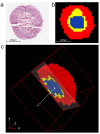
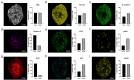
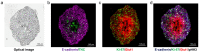
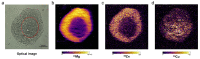
Mass spectrometry imaging (MSI) is an established analytical tool capable of defining and understanding complex tissues by determining the spatial distribution of biological molecules. Three-dimensional (3D) cell culture models mimic the pathophysiological environment of in vivo tumors and are rapidly emerging as a valuable research tool. Here, multimodal MSI techniques were employed to characterize a novel aggregated 3D lung adenocarcinoma model, developed by the group to mimic the in vivo tissue. Regions of tumor heterogeneity and the hypoxic microenvironment were observed based on the spatial distribution of a variety of endogenous molecules. Desorption electrospray ionization (DESI)-MSI defined regions of a hypoxic core and a proliferative outer layer from metabolite distribution. Targeted metabolites (e.g., lactate, glutamine, and citrate) were mapped to pathways of glycolysis and the TCA cycle demonstrating tumor metabolic behavior. The first application of imaging mass cytometry (IMC) with 3D cell culture enabled single-cell phenotyping at 1 μm spatial resolution. Protein markers of proliferation (Ki-67) and hypoxia (glucose transporter 1) defined metabolic signaling in the aggregoid model, which complemented the metabolite data. Laser ablation inductively coupled plasma (LA-ICP)-MSI analysis localized endogenous elements including magnesium and copper, further differentiating the hypoxia gradient and validating the protein expression. Obtaining a large amount of molecular information on a complementary nature enabled an in-depth understanding of the biological processes within the novel tumor model. Combining powerful imaging techniques to characterize the aggregated 3D culture highlighted a future methodology with potential applications in cancer research and drug development.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Swales J. G.; Hamm G.; Clench M. R.; Goodwin R. J. A. Mass spectrometry imaging and its application in pharmaceutical research and development: A concise review. Int. J. Mass Spectrom. 2019, 437, 99–112. 10.1016/j.ijms.2018.02.007. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

