Optimization of Acetazolamide-Based Scaffold as Potent Inhibitors of Vancomycin-Resistant Enterococcus
- PMID: 32787141
- PMCID: PMC8317130
- DOI: 10.1021/acs.jmedchem.0c00734
Optimization of Acetazolamide-Based Scaffold as Potent Inhibitors of Vancomycin-Resistant Enterococcus
Abstract
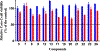
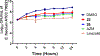
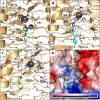
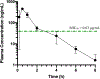
Vancomycin-resistant enterococci (VRE) are the second leading cause of hospital-acquired infections (HAIs) attributed to a drug-resistant bacterium in the United States, and resistance to the frontline treatments is well documented. To combat VRE, we have repurposed the FDA-approved carbonic anhydrase drug acetazolamide to design potent antienterococcal agents. Through structure-activity relationship optimization we have arrived at two leads possessing improved potency against clinical VRE strains from MIC = 2 μg/mL (acetazolamide) to MIC = 0.007 μg/mL (22) and 1 μg/mL (26). Physicochemical properties were modified to design leads that have either high oral bioavailability to treat systemic infections or low intestinal permeability to treat VRE infections in the gastrointestinal tract. Our data suggest the intracellular targets for the molecules are putative α-carbonic and γ-carbonic anhydrases, and homology modeling and molecular dynamics simulations were performed. Together, this study presents potential anti-VRE therapeutic options to provide alternatives for problematic VRE infections.
Figures
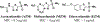
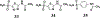



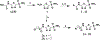
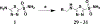
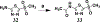
References
-
- Sievert DM; Ricks P; Edwards JR; Schneider A; Patel J; Srinivasan A; Kallen A; Limbago B; Fridkin S Antimicrobial-Resistant Pathogens Associated with Healthcare-Associated Infections Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control Hosp. Epidemiol. 2013, 34, 1–14. - PubMed
-
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2019; Atlanta, GA, 2019.
-
- Stagliano D; Susi A; Adams D; Nyland C Abstracts of Papers, Open Forum Infectious Diseases Fall 2017, Oct 7, 201; Epidemiology, Associated Conditions, and Outcomes of Hospital Associated Vancomycin-Resistant Enterococcus Infections in the US Military Health Care System. Infectious Disease Society of America: Arlington, VA, 2017.
-
- Rossolini GM; Arena F; Pecile P; Pollini S Update on the Antibiotic Resistance Crisis. Curr. Opin. Pharmacol. 2014, 18, 56–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Medical

