Caspases from scleractinian coral show unique regulatory features
- PMID: 32788218
- PMCID: PMC7586219
- DOI: 10.1074/jbc.RA120.014345
Caspases from scleractinian coral show unique regulatory features
Abstract
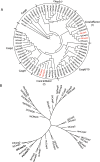
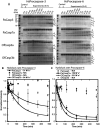
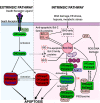
Coral reefs are experiencing precipitous declines around the globe with coral diseases and temperature-induced bleaching being primary drivers of these declines. Regulation of apoptotic cell death is an important component in the coral stress response. Although cnidaria are known to contain complex apoptotic signaling pathways, similar to those in vertebrates, the mechanisms leading to cell death are largely unexplored. We identified and characterized two caspases each from Orbicella faveolata, a disease-sensitive reef-building coral, and Porites astreoides, a disease-resistant reef-building coral. The caspases are predicted homologs of the human executioner caspases-3 and -7, but OfCasp3a (Orbicella faveolata caspase-3a) and PaCasp7a (Porites astreoides caspase-7a), which we show to be DXXDases, contain an N-terminal caspase activation/recruitment domain (CARD) similar to human initiator/inflammatory caspases. OfCasp3b (Orbicella faveolata caspase-3b) and PaCasp3 (Porites astreoides caspase-3), which we show to be VXXDases, have short pro-domains, like human executioner caspases. Our biochemical analyses suggest a mechanism in coral which differs from that of humans, where the CARD-containing DXXDase is activated on death platforms but the protease does not directly activate the VXXDase. The first X-ray crystal structure of a coral caspase, of PaCasp7a determined at 1.57 Å resolution, reveals a conserved fold and an N-terminal peptide bound near the active site that may serve as a regulatory exosite. The binding pocket has been observed in initiator caspases of other species. These results suggest mechanisms for the evolution of substrate selection while maintaining common activation mechanisms of CARD-mediated dimerization.
Keywords: CARD-caspase; allosteric regulation; apoptosis; caspase; coral apoptosis; coral immunity; cysteine protease; functional divergence; substrate selection; substrate specificity.
© 2020 Shrestha et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

