IL-6 regulates CCR5 expression and immunosuppressive capacity of MDSC in murine melanoma
- PMID: 32788238
- PMCID: PMC7422659
- DOI: 10.1136/jitc-2020-000949
IL-6 regulates CCR5 expression and immunosuppressive capacity of MDSC in murine melanoma
Abstract
Background: Myeloid-derived suppressor cells (MDSC) play a major role in the immunosuppressive melanoma microenvironment. They are generated under chronic inflammatory conditions characterized by the constant production of inflammatory cytokines, chemokines and growth factors, including IL-6. Recruitment of MDSC to the tumor is mediated by the interaction between chemokines and chemokine receptors, in particular C-C chemokine receptor (CCR)5. Here, we studied the mechanisms of CCR5 upregulation and increased immunosuppressive function of CCR5+ MDSC.
Methods: The immortalized myeloid suppressor cell line MSC-2, primary immature myeloid cells and in vitro differentiated MDSC were used to determine factors and molecular mechanisms regulating CCR5 expression and immunosuppressive markers at the mRNA and protein levels. The relevance of the identified pathways was validated on the RET transgenic mouse melanoma model, which was also used to target the identified pathways in vivo.
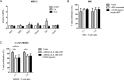
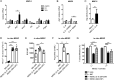
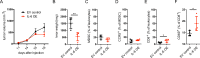
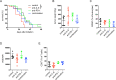
Results: IL-6 upregulated the expression of CCR5 and arginase 1 in MDSC by a STAT3-dependent mechanism. MDSC differentiated in the presence of IL-6 strongly inhibited CD8+ T cell functions compared with MDSC differentiated without IL-6. A correlation between IL-6 levels, phosphorylated STAT3 and CCR5 expression in tumor-infiltrating MDSC was demonstrated in the RET transgenic melanoma mouse model. Surprisingly, IL-6 overexpressing tumors grew significantly slower in mice accompanied by CD8+ T cell activation. Moreover, transgenic melanoma-bearing mice treated with IL-6 blocking antibodies showed significantly accelerated tumor development.
Conclusion: Our in vitro and ex vivo findings demonstrated that IL-6 induced CCR5 expression and a strong immunosuppressive activity of MDSC, highlighting this cytokine as a promising target for melanoma immunotherapy. However, IL-6 blocking therapy did not prove to be effective in RET transgenic melanoma-bearing mice but rather aggravated tumor progression. Further studies are needed to identify particular combination therapies, cancer entities or patient subsets to benefit from the anti-IL-6 treatment.
Keywords: cytokines; immune evasion; melanoma; myeloid-derived suppressor cells.
© Author(s) (or their employer(s)) 2020. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
