An ATP-dependent partner switch links flagellar C-ring assembly with gene expression
- PMID: 32788349
- PMCID: PMC7456076
- DOI: 10.1073/pnas.2006470117
An ATP-dependent partner switch links flagellar C-ring assembly with gene expression
Abstract
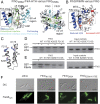
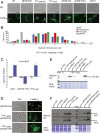
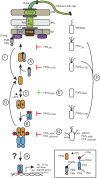
Bacterial flagella differ in their number and spatial arrangement. In many species, the MinD-type ATPase FlhG (also YlxH/FleN) is central to the numerical control of bacterial flagella, and its deletion in polarly flagellated bacteria typically leads to hyperflagellation. The molecular mechanism underlying this numerical control, however, remains enigmatic. Using the model species Shewanella putrefaciens, we show that FlhG links assembly of the flagellar C ring with the action of the master transcriptional regulator FlrA (named FleQ in other species). While FlrA and the flagellar C-ring protein FliM have an overlapping binding site on FlhG, their binding depends on the ATP-dependent dimerization state of FlhG. FliM interacts with FlhG independent of nucleotide binding, while FlrA exclusively interacts with the ATP-dependent FlhG dimer and stimulates FlhG ATPase activity. Our in vivo analysis of FlhG partner switching between FliM and FlrA reveals its mechanism in the numerical restriction of flagella, in which the transcriptional activity of FlrA is down-regulated through a negative feedback loop. Our study demonstrates another level of regulatory complexity underlying the spationumerical regulation of flagellar biogenesis and implies that flagellar assembly transcriptionally regulates the production of more initial building blocks.
Keywords: ATPase; flagellum; nanomachine; regulation; spatiotemporal.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

