Salmonella enterica serovar Typhi exposure elicits ex vivo cell-type-specific epigenetic changes in human gut cells
- PMID: 32788681
- PMCID: PMC7423951
- DOI: 10.1038/s41598-020-70492-2
Salmonella enterica serovar Typhi exposure elicits ex vivo cell-type-specific epigenetic changes in human gut cells
Erratum in
-
Author Correction: Salmonella enterica serovar Typhi exposure elicits ex vivo cell-type-specific epigenetic changes in human gut cells.Sci Rep. 2021 Jan 4;11(1):264. doi: 10.1038/s41598-020-79464-y. Sci Rep. 2021. PMID: 33398004 Free PMC article. No abstract available.
Abstract
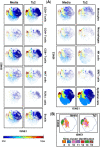
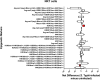
Salmonella enterica serovar Typhi (S. Typhi) causes substantial morbidity and mortality worldwide, particularly among young children. Humans develop an array of mucosal immune responses following S. Typhi infection. Whereas the cellular mechanisms involved in S. Typhi infection have been intensively studied, very little is known about the early chromatin modifications occurring in the human gut microenvironment that influence downstream immune responses. To address this gap in knowledge, cells isolated from human terminal ileum exposed ex vivo to the wild-type S. Typhi strain were stained with a 33-metal-labeled antibody panel for mass cytometry analyses of the early chromatin modifications modulated by S. Typhi. We measured the cellular levels of 6 classes of histone modifications, and 1 histone variant in 11 major cell subsets (i.e., B, CD3 + T, CD4 + T, CD8 + T, NK, TCR-γδ, Mucosal associated invariant (MAIT), and NKT cells as well as monocytes, macrophages, and epithelial cells). We found that arginine methylation might regulate the early-differentiation of effector-memory CD4+ T-cells following exposure to S. Typhi. We also found S. Typhi-induced post-translational modifications in histone methylation and acetylation associated with epithelial cells, NKT, MAIT, TCR-γδ, Monocytes, and CD8 + T-cells that are related to both gene activation and silencing.
Conflict of interest statement
The authors declare no competing interests.
Figures









References
-
- Levine, M. M. in Plotkin's vaccines (eds S A Plotkin, W A Oreinstein, P A Offit, & K M Edwards) Ch. 61, 1114–1144 (Elsevier, 2018).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

