Arginine as an environmental and metabolic cue for cyclic diguanylate signalling and biofilm formation in Pseudomonas putida
- PMID: 32788689
- PMCID: PMC7423604
- DOI: 10.1038/s41598-020-70675-x
Arginine as an environmental and metabolic cue for cyclic diguanylate signalling and biofilm formation in Pseudomonas putida
Abstract
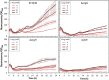
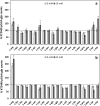
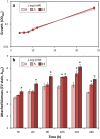
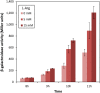
Cyclic diguanylate (c-di-GMP) is a broadly conserved intracellular second messenger that influences different bacterial processes, including virulence, stress tolerance or social behaviours and biofilm development. Although in most cases the environmental cue that initiates the signal transduction cascade leading to changes in cellular c-di-GMP levels remains unknown, certain L- and D-amino acids have been described to modulate c-di-GMP turnover in some bacteria. In this work, we have analysed the influence of L-amino acids on c-di-GMP levels in the plant-beneficial bacterium Pseudomonas putida KT2440, identifying L-arginine as the main one causing a significant increase in c-di-GMP. Both exogenous (environmental) and endogenous (biosynthetic) L-arginine influence biofilm formation by P. putida through changes in c-di-GMP content and altered expression of structural elements of the biofilm extracellular matrix. The contribution of periplasmic binding proteins forming part of amino acid transport systems to the response to environmental L-arginine was also studied. Contrary to what has been described in other bacteria, in P. putida these proteins seem not to be directly responsible for signal transduction. Rather, their contribution to global L-arginine pools appears to determine changes in c-di-GMP turnover. We propose that arginine plays a connecting role between cellular metabolism and c-di-GMP signalling in P. putida.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Jenal U, Reinders A, Lori C. Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 2017;15:271–284. - PubMed
-
- Matilla MA, Travieso ML, Ramos JL, Ramos-González MI. Cyclic diguanylate turnover mediated by the sole GGDEF/EAL response regulator in Pseudomonas putida: Its role in the rhizosphere and an analysis of its target processes. Environ. Microbiol. 2011;13:1745–1766. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

