cDC1 prime and are licensed by CD4+ T cells to induce anti-tumour immunity
- PMID: 32788723
- PMCID: PMC7469755
- DOI: 10.1038/s41586-020-2611-3
cDC1 prime and are licensed by CD4+ T cells to induce anti-tumour immunity
Abstract
Conventional type 1 dendritic cells (cDC1)1 are thought to perform antigen cross-presentation, which is required to prime CD8+ T cells2,3, whereas cDC2 are specialized for priming CD4+ T cells4,5. CD4+ T cells are also considered to help CD8+ T cell responses through a variety of mechanisms6-11, including a process whereby CD4+ T cells 'license' cDC1 for CD8+ T cell priming12. However, this model has not been directly tested in vivo or in the setting of help-dependent tumour rejection. Here we generated an Xcr1Cre mouse strain to evaluate the cellular interactions that mediate tumour rejection in a model requiring CD4+ and CD8+ T cells. As expected, tumour rejection required cDC1 and CD8+ T cell priming required the expression of major histocompatibility class I molecules by cDC1. Unexpectedly, early priming of CD4+ T cells against tumour-derived antigens also required cDC1, and this was not simply because they transport antigens to lymph nodes for processing by cDC2, as selective deletion of major histocompatibility class II molecules in cDC1 also prevented early CD4+ T cell priming. Furthermore, deletion of either major histocompatibility class II or CD40 in cDC1 impaired tumour rejection, consistent with a role for cognate CD4+ T cell interactions and CD40 signalling in cDC1 licensing. Finally, CD40 signalling in cDC1 was critical not only for CD8+ T cell priming, but also for initial CD4+ T cell activation. Thus, in the setting of tumour-derived antigens, cDC1 function as an autonomous platform capable of antigen processing and priming for both CD4+ and CD8+ T cells and of the direct orchestration of their cross-talk that is required for optimal anti-tumour immunity.
Figures
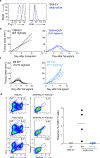
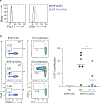


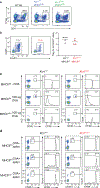








Comment in
-
A dendritic cell multitasks to tackle cancer.Nature. 2020 Aug;584(7822):533-534. doi: 10.1038/d41586-020-02339-9. Nature. 2020. PMID: 32788699 No abstract available.
References
-
- Dudziak D et al. Differential antigen processing by dendritic cell subsets in vivo. Science 315, 107–111 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA190700/CA/NCI NIH HHS/United States
- R01 AI129545/AI/NIAID NIH HHS/United States
- T32 CA009573/CA/NCI NIH HHS/United States
- F30 DK112466/DK/NIDDK NIH HHS/United States
- R01 NS106289/NS/NINDS NIH HHS/United States
- T32 AI007163/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- UL1 TR002345/TR/NCATS NIH HHS/United States
- T32 CA009621/CA/NCI NIH HHS/United States
- R01 AI150297/AI/NIAID NIH HHS/United States
- R01 CA248919/CA/NCI NIH HHS/United States
- P30 AR073752/AR/NIAMS NIH HHS/United States
- R01 AI162643/AI/NIAID NIH HHS/United States
- F30 DK108498/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

