An approach for normalization and quality control for NanoString RNA expression data
- PMID: 32789507
- PMCID: PMC8138885
- DOI: 10.1093/bib/bbaa163
An approach for normalization and quality control for NanoString RNA expression data
Abstract
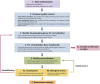
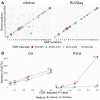
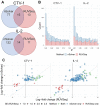
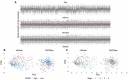
The NanoString RNA counting assay for formalin-fixed paraffin embedded samples is unique in its sensitivity, technical reproducibility and robustness for analysis of clinical and archival samples. While commercial normalization methods are provided by NanoString, they are not optimal for all settings, particularly when samples exhibit strong technical or biological variation or where housekeeping genes have variable performance across the cohort. Here, we develop and evaluate a more comprehensive normalization procedure for NanoString data with steps for quality control, selection of housekeeping targets, normalization and iterative data visualization and biological validation. The approach was evaluated using a large cohort ($N=\kern0.5em 1649$) from the Carolina Breast Cancer Study, two cohorts of moderate sample size ($N=359$ and$130$) and a small published dataset ($N=12$). The iterative process developed here eliminates technical variation (e.g. from different study phases or sites) more reliably than the three other methods, including NanoString's commercial package, without diminishing biological variation, especially in long-term longitudinal multiphase or multisite cohorts. We also find that probe sets validated for nCounter, such as the PAM50 gene signature, are impervious to batch issues. This work emphasizes that systematic quality control, normalization and visualization of NanoString nCounter data are an imperative component of study design that influences results in downstream analyses.
Keywords: NanoString nCounter expression; data visualization; gene expression normalization; quality control.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Geiss GK, Bumgarner RE, Birditt B, et al. . Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008;26:317–25. - PubMed
-
- Veldman-Jones MH, Brant R, Rooney C, et al. . Evaluating robustness and sensitivity of the NanoString technologies nCounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Res 2015;75:2587–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

