ADD Force Field for Sugars and Polyols: Predicting the Additivity of Protein-Osmolyte Interaction
- PMID: 32790371
- PMCID: PMC7901642
- DOI: 10.1021/acs.jpcb.0c05345
ADD Force Field for Sugars and Polyols: Predicting the Additivity of Protein-Osmolyte Interaction
Abstract
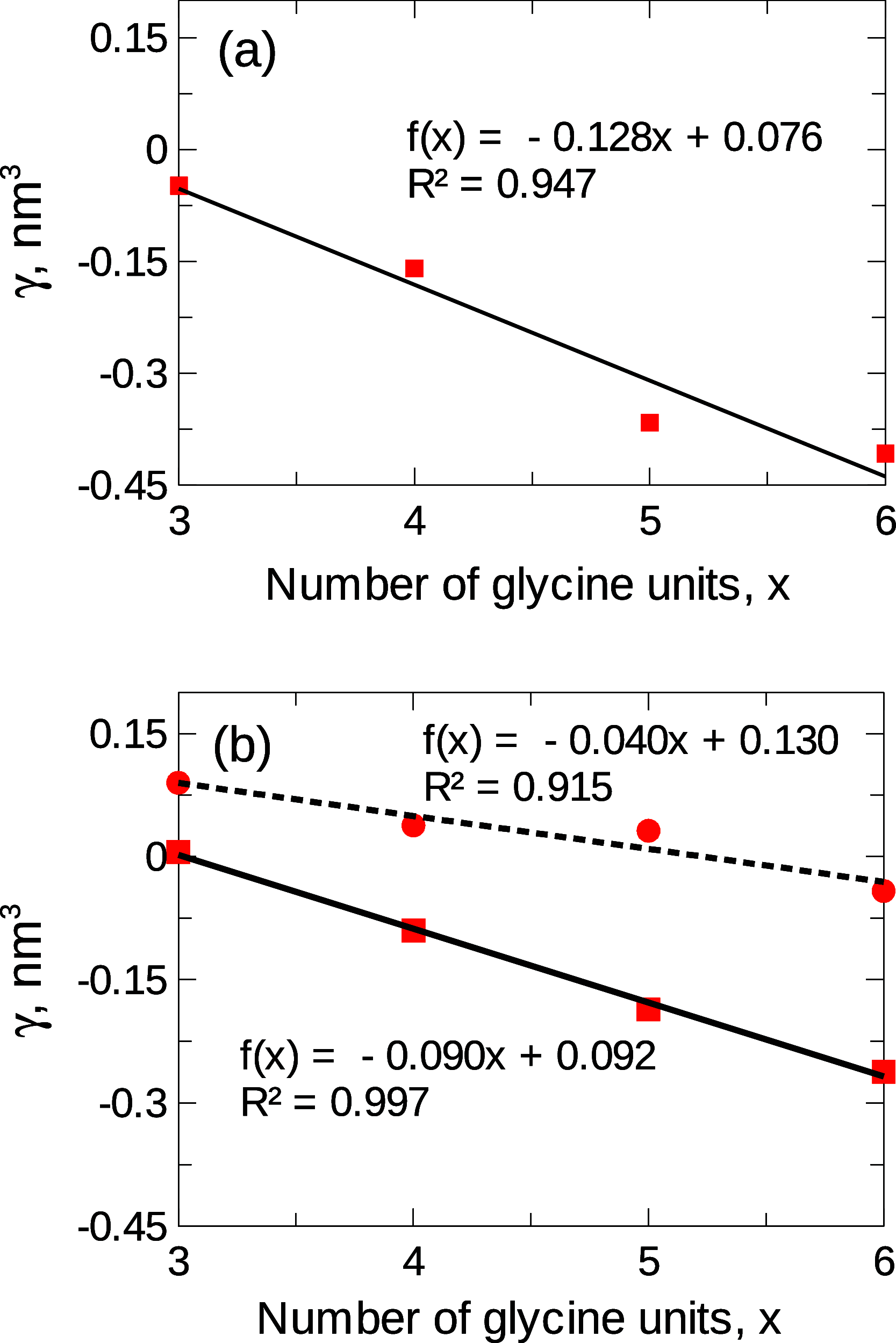
The protein-osmolyte interaction has been shown experimentally to follow an additive construct, where the individual osmolyte-backbone and osmolyte-side-chain interactions contribute to the overall conformational stability of proteins. Here, we computationally reconstruct this additive relation using molecular dynamics simulations, focusing on sugars and polyols, including sucrose and sorbitol, as model osmolytes. A new set of parameters (ADD) is developed for this purpose, using the individual Kirkwood-Buff integrals for sugar-backbone and sugar-side-chain interactions as target experimental data. We show that the ADD parameters can reproduce the additivity of protein-sugar interactions and correctly predict sucrose and sorbitol self-association and their interaction with water. The accurate description of the separate osmolyte-backbone and osmolyte-side-chain contributions also automatically translates into a good prediction of preferential exclusion from the surface of ribonuclease A and α-chymotrypsinogen A. The description of sugar polarity is improved compared to previous force fields, resulting in closer agreement with the experimental data and better compatibility with charged groups, such as the guanidinium moiety. The ADD parameters are developed in combination with the CHARMM36m force field for proteins, but good compatibility is also observed with the AMBER 99SB-ILDN and the OPLS-AA force fields. Overall, exploiting the additivity of protein-osmolyte interactions is a promising approach for the development of new force fields.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
-
- Lee J. C.; Timasheff S. N. The stabilization of proteins by sucrose. J. Biol. Chem. 1981, 256, 7193–7201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

