Does SARS-CoV-2 Bind to Human ACE2 More Strongly Than Does SARS-CoV?
- PMID: 32790406
- PMCID: PMC7433338
- DOI: 10.1021/acs.jpcb.0c04511
Does SARS-CoV-2 Bind to Human ACE2 More Strongly Than Does SARS-CoV?
Abstract
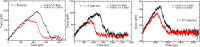
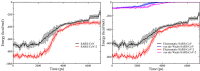
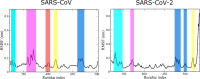
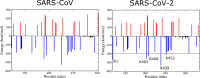
The 2019 novel coronavirus (SARS-CoV-2) epidemic, which was first reported in December 2019 in Wuhan, China, was declared a pandemic by the World Health Organization in March 2020. Genetically, SARS-CoV-2 is closely related to SARS-CoV, which caused a global epidemic with 8096 confirmed cases in more than 25 countries from 2002 to 2003. Given the significant morbidity and mortality rate, the current pandemic poses a danger to all of humanity, prompting us to understand the activity of SARS-CoV-2 at the atomic level. Experimental studies have revealed that spike proteins of both SARS-CoV-2 and SARS-CoV bind to angiotensin-converting enzyme 2 (ACE2) before entering the cell for replication. However, the binding affinities reported by different groups seem to contradict each other. Wrapp et al. (Science 2020, 367, 1260-1263) showed that the spike protein of SARS-CoV-2 binds to the ACE2 peptidase domain (ACE2-PD) more strongly than does SARS-CoV, and this fact may be associated with a greater severity of the new virus. However, Walls et al. (Cell 2020, 181, 281-292) reported that SARS-CoV-2 exhibits a higher binding affinity, but the difference between the two variants is relatively small. To understand the binding mechnism and experimental results, we investigated how the receptor binding domain (RBD) of SARS-CoV (SARS-CoV-RBD) and SARS-CoV-2 (SARS-CoV-2-RBD) interacts with a human ACE2-PD using molecular modeling. We applied a coarse-grained model to calculate the dissociation constant and found that SARS-CoV-2 displays a 2-fold higher binding affinity. Using steered all-atom molecular dynamics simulations, we demonstrate that, like a coarse-grained simulation, SARS-CoV-2-RBD was associated with ACE2-PD more strongly than was SARS-CoV-RBD, as evidenced by a higher rupture force and larger pulling work. We show that the binding affinity of both viruses to ACE2 is driven by electrostatic interactions.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Organization, W. H. WHO Director-General’s opening remarks at the media briefing on COVID-19–11 March 2020. Geneva, Switzerland, 2020.
-
- Chan J. F.-W.; Kok K.-H.; Zhu Z.; Chu H.; To K. K.-W.; Yuan S.; Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes Infect. 2020, 9, 221–236. 10.1080/22221751.2020.1719902. - DOI - PMC - PubMed
-
- Gorbalenya A. E.; Baker S. C.; Baric R. S.; de Groot R. J.; Drosten C.; Gulyaeva A. A.; Haagmans B. L.; Lauber C.; Leontovich A. M.; Neuman B. W.; et al. Coronaviridae Study Group of the International Committee on Taxonomy of, V. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. 10.1038/s41564-020-0695-z. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

