Human Golgi phosphoprotein 3 is an effector of RAB1A and RAB1B
- PMID: 32790781
- PMCID: PMC7425898
- DOI: 10.1371/journal.pone.0237514
Human Golgi phosphoprotein 3 is an effector of RAB1A and RAB1B
Abstract
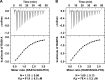
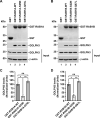
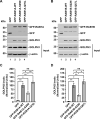
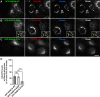
Golgi phosphoprotein 3 (GOLPH3) is a peripheral membrane protein localized at the trans-Golgi network that is also distributed in a large cytosolic pool. GOLPH3 has been involved in several post-Golgi protein trafficking events, but its precise function at the molecular level is not well understood. GOLPH3 is also considered the first oncoprotein of the Golgi apparatus, with important roles in several types of cancer. Yet, it is unknown how GOLPH3 is regulated to achieve its contribution in the mechanisms that lead to tumorigenesis. Binding of GOLPH3 to Golgi membranes depends on its interaction to phosphatidylinositol-4-phosphate. However, an early finding showed that GTP promotes the binding of GOLPH3 to Golgi membranes and vesicles. Nevertheless, it remains largely unknown whether this response is consequence of the function of GTP-dependent regulatory factors, such as proteins of the RAB family of small GTPases. Interestingly, in Drosophila melanogaster the ortholog of GOLPH3 interacts with- and behaves as effector of the ortholog of RAB1. However, there is no experimental evidence implicating GOLPH3 as a possible RAB1 effector in mammalian cells. Here, we show that human GOLPH3 interacted directly with either RAB1A or RAB1B, the two isoforms of RAB1 in humans. The interaction was nucleotide dependent and it was favored with GTP-locked active state variants of these GTPases, indicating that human GOLPH3 is a bona fide effector of RAB1A and RAB1B. Moreover, the expression in cultured cells of the GTP-locked variants resulted in less distribution of GOLPH3 in the Golgi apparatus, suggesting an intriguing model of GOLPH3 regulation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Wu CC, Taylor RS, Lane DR, Ladinsky MS, Weisz JA, Howell KE. GMx33: a novel family of trans-Golgi proteins identified by proteomics. Traffic. 2000;1(12):963–75. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

