Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses
- PMID: 32791036
- PMCID: PMC7402670
- DOI: 10.1016/j.immuni.2020.07.026
Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses
Abstract
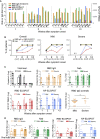
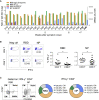
The SARS-CoV-2 pandemic has resulted in millions of infections, yet the role of host immune responses in early COVID-19 pathogenesis remains unclear. By investigating 17 acute and 24 convalescent patients, we found that acute SARS-CoV-2 infection resulted in broad immune cell reduction including T, natural killer, monocyte, and dendritic cells (DCs). DCs were significantly reduced with functional impairment, and ratios of conventional DCs to plasmacytoid DCs were increased among acute severe patients. Besides lymphocytopenia, although neutralizing antibodies were rapidly and abundantly generated in patients, there were delayed receptor binding domain (RBD)- and nucleocapsid protein (NP)-specific T cell responses during the first 3 weeks after symptoms onset. Moreover, acute RBD- and NP-specific T cell responses included relatively more CD4 T cells than CD8 T cells. Our findings provided evidence that impaired DCs, together with timely inverted strong antibody but weak CD8 T cell responses, could contribute to acute COVID-19 pathogenesis and have implications for vaccine development.
Keywords: COVID-19; SARS-CoV-2; T cell immune response; acute infection; convalescent; dendritic cell; neutralizing antibody; nucleocapsid protein; receptor-binding domain.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures




Similar articles
-
Long-term infection of SARS-CoV-2 changed the body's immune status.Clin Immunol. 2020 Sep;218:108524. doi: 10.1016/j.clim.2020.108524. Epub 2020 Jul 11. Clin Immunol. 2020. PMID: 32659373 Free PMC article.
-
Immune characteristics of severe and critical COVID-19 patients.Signal Transduct Target Ther. 2020 Aug 31;5(1):179. doi: 10.1038/s41392-020-00296-3. Signal Transduct Target Ther. 2020. PMID: 32868756 Free PMC article. No abstract available.
-
Single-cell landscape of immunological responses in patients with COVID-19.Nat Immunol. 2020 Sep;21(9):1107-1118. doi: 10.1038/s41590-020-0762-x. Epub 2020 Aug 12. Nat Immunol. 2020. PMID: 32788748
-
Convalescent donor SARS-COV-2-specific cytotoxic T lymphocyte infusion as a possible treatment option for COVID-19 patients with severe disease has not received enough attention till date.Br J Haematol. 2020 Jun;189(6):1062-1063. doi: 10.1111/bjh.16780. Epub 2020 May 19. Br J Haematol. 2020. PMID: 32369628 Review. No abstract available.
-
On the genetics and immunopathogenesis of COVID-19.Clin Immunol. 2020 Nov;220:108591. doi: 10.1016/j.clim.2020.108591. Epub 2020 Sep 10. Clin Immunol. 2020. PMID: 32920210 Free PMC article. Review.
Cited by
-
Combined plasma levels of IL-10 and testosterone, but not soluble HLA-G5, predict the risk of death in COVID-19 patients.Andrology. 2023 Jan;11(1):32-44. doi: 10.1111/andr.13334. Epub 2022 Nov 21. Andrology. 2023. PMID: 36323494 Free PMC article.
-
Infection and Immune Memory: Variables in Robust Protection by Vaccines Against SARS-CoV-2.Front Immunol. 2021 May 11;12:660019. doi: 10.3389/fimmu.2021.660019. eCollection 2021. Front Immunol. 2021. PMID: 34046033 Free PMC article. Review.
-
Mitochondrial regulation of acute extrafollicular B-cell responses to COVID-19 severity.Clin Transl Med. 2022 Sep;12(9):e1025. doi: 10.1002/ctm2.1025. Clin Transl Med. 2022. PMID: 36103567 Free PMC article.
-
An Impaired Inflammatory and Innate Immune Response in COVID-19.Mol Cells. 2021 Jun 30;44(6):384-391. doi: 10.14348/molcells.2021.0068. Mol Cells. 2021. PMID: 34098591 Free PMC article. Review.
-
The serum metabolome of COVID-19 patients is distinctive and predictive.Metabolism. 2021 May;118:154739. doi: 10.1016/j.metabol.2021.154739. Epub 2021 Mar 2. Metabolism. 2021. PMID: 33662365 Free PMC article.
References
-
- Chan J.F., Yip C.C., To K.K., Tang T.H., Wong S.C., Leung K.H., Fung A.Y., Ng A.C., Zou Z., Tsoi H.W. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J. Clin. Microbiol. 2020;58:e00310–e00320. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

