Oral Microbiome Geography: Micron-Scale Habitat and Niche
- PMID: 32791109
- PMCID: PMC7604680
- DOI: 10.1016/j.chom.2020.07.009
Oral Microbiome Geography: Micron-Scale Habitat and Niche
Abstract
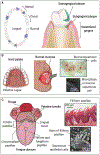
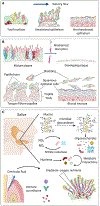
The mouth presents a multiplicity of local environments in communication with one another via saliva. The spatial organization of microbes within the mouth is shaped by opposing forces in dynamic equilibrium-salivary flow and adhesion, shedding and colonization-and by interactions among and between microbes and the host. Here we review recent evidence confirming that oral microbes are specialized for individual habitats within the mouth and that microbial habitats and niches are defined by micron-scale gradients in combination with short- and long-range interactions. Micron-scale structure illuminates the roles of individual taxa and provides insight into their community ecology and potential pathogenicity.
Copyright © 2020. Published by Elsevier Inc.
Figures

References
-
- Datar U, Angadi PV, Hallikerimath S, and Kale AD (2013). Cytological assessment of Barr bodies using aceto-orcein and papanicolaou stains in buccal mucosal smears and their sex estimation efficacy in an Indian sample. Acta Cytol. 57, 516–521. - PubMed
-
- Dawes C (2003). Estimates, from salivary analyses, of the turnover time of the oral mucosal epithelium in humans and the number of bacteria in an edentulous mouth. Arch. Oral Biol 48, 329–336. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

