The Skin Microbiota: Balancing Risk and Reward
- PMID: 32791112
- PMCID: PMC7444652
- DOI: 10.1016/j.chom.2020.06.017
The Skin Microbiota: Balancing Risk and Reward
Abstract
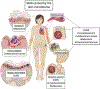
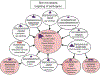
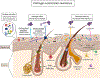
The skin microbiome is an ecosystem comprised of a multitude of microbial species interacting with their surroundings, including other microbes and host epithelial and immune cells. These interactions are the basis of important roles within the skin microbiome that provide benefit to the host, boosting multiple aspects of barrier function, a critical function of this essential organ. However, with reward always comes risk; resident skin microbes function in a context-dependent manner, set on the backdrop of a dynamic host and microbial milieu. Here, we discuss the reward of hosting a microbial ecosystem on the skin, including protection from pathogens and tuning of the skin microenvironment. We also give consideration to how these skin residents, often termed "commensals" can cause disorder, damage, and promote skin disease.
Keywords: commensal microbes; cutaneous disease; dermatology; microbiome; skin.
Copyright © 2020 Elsevier Inc. All rights reserved.
Figures

References
-
- Bawdon D, Cox DS, Ashford D, James AG, and Thomas GH (2015). Identification of axillary Staphylococcus sp. involved in the production of the malodorous thioalcohol 3-methyl-3-sufanylhexan-1-ol. FEMS Microbiol. Lett. 362, fnv111. - PubMed
-
- Brown SK, and Shalita AR (1998). Acne vulgaris. Lancet 351, 1871–1876. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

