Proteomic analysis revealed common, unique and systemic signatures in gender-dependent hepatocarcinogenesis
- PMID: 32792008
- PMCID: PMC7427087
- DOI: 10.1186/s13293-020-00316-5
Proteomic analysis revealed common, unique and systemic signatures in gender-dependent hepatocarcinogenesis
Abstract
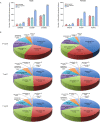
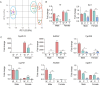
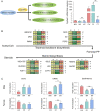
Hepatocellular carcinoma (HCC) is the most common liver cancer and is highly malignant. Male prevalence and frequent activation of the Ras signaling pathway are distinct characteristics of HCC. However, the underlying mechanisms remain to be elucidated. By exploring Hras12V transgenic mice showing male-biased hepatocarcinogenesis, we performed a high-throughput comparative proteomic analysis based on tandem-mass-tag (TMT) labeling combined with liquid chromatography-tandem mass spectrometry (LC-MS/MS) on the tissue samples obtained from HCC (T) and their paired adjacent precancerous (P) of Hras12V transgenic male and female mice (Ras-Tg) and normal liver (W) of wild-type male and female mice (Non-Tg). The further validation and investigation were performed using quantitative real-time PCR and western blot. Totally, 5193 proteins were quantified, originating from 5733 identified proteins. Finally, 1344 differentially expressed proteins (DEPs) (quantified in all examined samples; |ratios| ≥ 1.5, p < 0.05) were selected for further analysis. Comparison within W, P, and T of males and females indicated that the number of DEPs in males was much higher than that in females. Bioinformatics analyses showed the common and unique cluster-enriched items between sexes, indicating the common and gender-disparate pathways towards HCC. Expression change pattern analysis revealed HCC positive/negative-correlated and ras oncogene positive/negative-correlated DEPs and pathways. In addition, it showed that the ras oncogene gradually and significantly reduced the responses to sex hormones from hepatocytes to hepatoma cells and therefore shrunk the gender disparity between males and females, which may contribute to the cause of the loss of HCC clinical responses to the therapeutic approaches targeting sex hormone pathways. Additionally, gender disparity in the expression levels of key enzymes involved in retinol metabolism and terpenoid backbone/steroid biosynthesis pathways may contribute to male prevalence in hepatocarcinogenesis. Further, the biomarkers, SAA2, Orm2, and Serpina1e, may be sex differences. In conclusion, common and unique DEPs and pathways toward HCC initiated by ras oncogene from sexually dimorphic hepatocytes provide valuable and novel insights into clinical investigation and practice.
Keywords: Gender disparity; Hepatocellular carcinoma; Proteomics; Ras oncogene; Tandem-mass-tag (TMT).
Conflict of interest statement
The authors declared that they have no conflicts of interest to this work.
Figures




Similar articles
-
Omics-Based Identification of Shared and Gender Disparity Routes in Hras12V-Induced Hepatocarcinogenesis: An Important Role for Dlk1-Dio3 Genomic Imprinting Region.Front Genet. 2021 May 31;12:620594. doi: 10.3389/fgene.2021.620594. eCollection 2021. Front Genet. 2021. PMID: 34135934 Free PMC article.
-
Transcriptomic landscape of Hras12V oncogene-induced hepatocarcinogenesis with gender disparity.BMC Cancer. 2025 Jan 16;25(1):94. doi: 10.1186/s12885-025-13476-7. BMC Cancer. 2025. PMID: 39819515 Free PMC article.
-
Differential Proteomic Analysis of Gender-dependent Hepatic Tumorigenesis in Hras12V Transgenic Mice.Mol Cell Proteomics. 2017 Aug;16(8):1475-1490. doi: 10.1074/mcp.M116.065474. Epub 2017 May 16. Mol Cell Proteomics. 2017. PMID: 28512230 Free PMC article.
-
Exploiting gender-based biomarkers and drug targets: advancing personalized therapeutic strategies in hepatocellular carcinoma.Front Pharmacol. 2024 Jun 20;15:1433540. doi: 10.3389/fphar.2024.1433540. eCollection 2024. Front Pharmacol. 2024. PMID: 38966543 Free PMC article. Review.
-
Divergent impact of gender in advancement of liver injuries, diseases, and carcinogenesis.Front Biosci (Schol Ed). 2018 Jan 1;10(1):65-100. doi: 10.2741/s501. Front Biosci (Schol Ed). 2018. PMID: 28930519 Review.
Cited by
-
Omics-Based Identification of Shared and Gender Disparity Routes in Hras12V-Induced Hepatocarcinogenesis: An Important Role for Dlk1-Dio3 Genomic Imprinting Region.Front Genet. 2021 May 31;12:620594. doi: 10.3389/fgene.2021.620594. eCollection 2021. Front Genet. 2021. PMID: 34135934 Free PMC article.
-
The Niemann-Pick C1 Protein of Patients with Hepatocellular Carcinoma Is Associated with Survival Time in Males and Tumor Size in Females.Biomedicines. 2025 Jul 13;13(7):1707. doi: 10.3390/biomedicines13071707. Biomedicines. 2025. PMID: 40722778 Free PMC article.
-
Hepatic Bone Morphogenetic Protein and Activin Membrane-Bound Inhibitor Levels Decline in Hepatitis C but Are Not Associated with Progression of Hepatocellular Carcinoma.Biomedicines. 2024 Oct 19;12(10):2397. doi: 10.3390/biomedicines12102397. Biomedicines. 2024. PMID: 39457709 Free PMC article.
-
Quantitative proteomic profiling reveals sexual dimorphism in the retina and RPE of C57BL6 mice.Biol Sex Differ. 2024 Oct 30;15(1):87. doi: 10.1186/s13293-024-00645-9. Biol Sex Differ. 2024. PMID: 39478535 Free PMC article.
-
Transcriptomic landscape of Hras12V oncogene-induced hepatocarcinogenesis with gender disparity.BMC Cancer. 2025 Jan 16;25(1):94. doi: 10.1186/s12885-025-13476-7. BMC Cancer. 2025. PMID: 39819515 Free PMC article.
References
-
- SH TK, Hashizume M, Hirohata T. Serum testosterone estradiol ratio and the development of hepatocellular carcinoma among male cirrhotic patients. Cancer Res. 2000;60:18:5106–5110. - PubMed
-
- Wang AG, Moon HB, Lee MR, Hwang CY, Kwon KS, Yu SL, et al. Gender-dependent hepatic alterations in H-ras12V transgenic mice. J Hepatol 2005;43:5:836-844; doi:10.1016/j.jhep.2005.04.012. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

