Identification of Undetected Monogenic Cardiovascular Disorders
- PMID: 32792077
- PMCID: PMC7428466
- DOI: 10.1016/j.jacc.2020.06.037
Identification of Undetected Monogenic Cardiovascular Disorders
Abstract
Background: Monogenic diseases are individually rare but collectively common, and are likely underdiagnosed.
Objectives: The purpose of this study was to estimate the prevalence of monogenic cardiovascular diseases (MCVDs) and potentially missed diagnoses in a cardiovascular cohort.
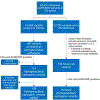
Methods: Exomes from 8,574 individuals referred for cardiac catheterization were analyzed. Pathogenic/likely pathogenic (P/LP) variants associated with MCVD (cardiomyopathies, arrhythmias, connective tissue disorders, and familial hypercholesterolemia were identified. Electronic health records (EHRs) were reviewed for individuals harboring P/LP variants who were predicted to develop disease (G+). G+ individuals who did not have a documented relevant diagnosis were classified into groups of whether they may represent missed diagnoses (unknown, unlikely, possible, probable, or definite) based on relevant diagnostic criteria/features for that disease.
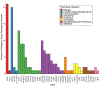
Results: In total, 159 P/LP variants were identified; 2,361 individuals harbored at least 1 P/LP variant, of whom 389 G+ individuals (4.5% of total cohort) were predicted to have at least 1 MCVD. EHR review of 342 G+ individuals predicted to have 1 MCVD with sufficient EHR data revealed that 52 had been given the relevant clinical diagnosis. The remaining 290 individuals were classified as potentially having an MCVD as follows: 193 unlikely (66.6%), 50 possible (17.2%), 30 probable (10.3%), and 17 definite (5.9%). Grouping possible, probable, definite, and known diagnoses, 149 were considered to have an MCVD. Novel MCVD pathogenic variants were identified in 16 individuals.
Conclusions: Overall, 149 individuals (1.7% of cohort) had MCVDs, but only 35% were diagnosed. These patients represents a "missed opportunity," which could be addressed by greater use of genetic testing of patients seen by cardiologists.
Keywords: amyloidosis; exome sequencing; genetics; monogenic diseases.
Copyright © 2020 American College of Cardiology Foundation. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures


Comment in
-
Testing for Not so Rare Monogenic Cardiovascular Diseases.J Am Coll Cardiol. 2020 Aug 18;76(7):809-811. doi: 10.1016/j.jacc.2020.06.060. J Am Coll Cardiol. 2020. PMID: 32792078 No abstract available.
References
-
- Luirink IK, Wiegman A, Kusters DM et al. 20-Year Follow-up of Statins in Children with Familial Hypercholesterolemia. N Engl J Med 2019;381:1547–1556. - PubMed
-
- Teekakirikul P, Kelly MA, Rehm HL, Lakdawala NK, Funke BH. Inherited cardiomyopathies: molecular genetics and clinical genetic testing in the postgenomic era. J Mol Diagn 2013;15:158–70. - PubMed
-
- Paterick TE, Humphries JA, Ammar KA et al. Aortopathies: etiologies, genetics, differential diagnosis, prognosis and management. Am J Med 2013;126:670–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

