The impact of transposable elements on tomato diversity
- PMID: 32792480
- PMCID: PMC7426864
- DOI: 10.1038/s41467-020-17874-2
The impact of transposable elements on tomato diversity
Erratum in
-
Author Correction: The impact of transposable elements on tomato diversity.Nat Commun. 2021 May 21;12(1):3203. doi: 10.1038/s41467-021-23578-y. Nat Commun. 2021. PMID: 34021160 Free PMC article. No abstract available.
Abstract
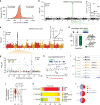
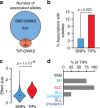
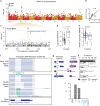
Tomatoes come in a multitude of shapes and flavors despite a narrow genetic pool. Here, we leverage whole-genome resequencing data available for 602 cultivated and wild accessions to determine the contribution of transposable elements (TEs) to tomato diversity. We identify 6,906 TE insertions polymorphisms (TIPs), which result from the mobilization of 337 distinct TE families. Most TIPs are low frequency variants and TIPs are disproportionately located within or adjacent to genes involved in environmental responses. In addition, genic TE insertions tend to have strong transcriptional effects and they can notably lead to the generation of multiple transcript isoforms. Using genome-wide association studies (GWAS), we identify at least 40 TIPs robustly associated with extreme variation in major agronomic traits or secondary metabolites and in most cases, no SNP tags the TE insertion allele. Collectively, these findings highlight the unique role of TE mobilization in tomato diversification, with important implications for breeding.
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
Mobile Transposable Elements Shape Plant Genome Diversity.Trends Plant Sci. 2020 Nov;25(11):1062-1064. doi: 10.1016/j.tplants.2020.08.003. Epub 2020 Aug 28. Trends Plant Sci. 2020. PMID: 32863103
References
-
- Lin T, et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014;46:1220–1226. - PubMed
-
- Gao L, et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 2019;51:1044–1051. - PubMed
-
- Huang X, Han B. Natural variations and genome-wide association studies in crop plants. Annu. Rev. Plant Biol. 2014;65:531–551. - PubMed
-
- Eathington SR, Crosbie TM, Edwards MD, Reiter RS, Bull JK. Molecular markers in a commercial breeding program. Crop Sci. 2007;47:S–154.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

