Neuronal subclass-selective proteomic analysis in Caenorhabditis elegans
- PMID: 32792517
- PMCID: PMC7426821
- DOI: 10.1038/s41598-020-70692-w
Neuronal subclass-selective proteomic analysis in Caenorhabditis elegans
Abstract
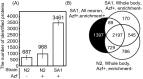
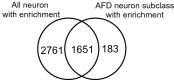
Neurons are categorised into many subclasses, and each subclass displays different morphology, expression patterns, connectivity and function. Changes in protein synthesis are critical for neuronal function. Therefore, analysing protein expression patterns in individual neuronal subclass will elucidate molecular mechanisms for memory and other functions. In this study, we used neuronal subclass-selective proteomic analysis with cell-selective bio-orthogonal non-canonical amino acid tagging. We selected Caenorhabditis elegans as a model organism because it shows diverse neuronal functions and simple neural circuitry. We performed proteomic analysis of all neurons or AFD subclass neurons that regulate thermotaxis in C. elegans. Mutant phenylalanyl tRNA synthetase (MuPheRS) was selectively expressed in all neurons or AFD subclass neurons, and azido-phenylalanine was incorporated into proteins in cells of interest. Azide-labelled proteins were enriched and proteomic analysis was performed. We identified 4,412 and 1,834 proteins from strains producing MuPheRS in all neurons and AFD subclass neurons, respectively. F23B2.10 (RING-type domain-containing protein) was identified only in neuronal cell-enriched proteomic analysis. We expressed GFP under the control of the 5' regulatory region of F23B2.10 and found GFP expression in neurons. We expect that more single-neuron specific proteomic data will clarify how protein composition and abundance affect characteristics of neuronal subclasses.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007;8:427–437. - PubMed
-
- Komatsu H, Mori I, Rhee J-S, Akaike N, Ohshima Y. Mutations in a cyclic nucleotide—gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron. 1996;17:707–718. - PubMed
-
- Hobert O, et al. Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron. 1997;19:345–357. - PubMed
-
- Kobayashi K, et al. Single-cell memory regulates a neural circuit for sensory behavior. Cell Rep. 2016;14:11–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

