Molecular evolution of coxsackievirus A24v in Cuba over 23-years, 1986-2009
- PMID: 32792520
- PMCID: PMC7427094
- DOI: 10.1038/s41598-020-70436-w
Molecular evolution of coxsackievirus A24v in Cuba over 23-years, 1986-2009
Abstract
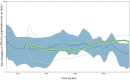
Coxsackievirus A24 variant (CVA24v) is a major causative agent of acute hemorrhagic conjunctivitis outbreaks worldwide, yet the evolutionary and transmission dynamics of the virus remain unclear. To address this, we analyzed and compared the 3C and partial VP1 gene regions of CVA24v isolates obtained from five outbreaks in Cuba between 1986 and 2009 and strains isolated worldwide. Here we show that Cuban strains were homologous to those isolated in Africa, the Americas and Asia during the same time period. Two genotypes of CVA24v (GIII and GIV) were repeatedly introduced into Cuba and they arose about two years before the epidemic was detected. The two genotypes co-evolved with a population size that is stable over time. However, nucleotide substitution rates peaked during pandemics with 4.39 × 10-3 and 5.80 × 10-3 substitutions per site per year for the 3C and VP1 region, respectively. The phylogeographic analysis identified 25 and 19 viral transmission routes based on 3C and VP1 regions, respectively. Pandemic viruses usually originated in Asia, and both China and Brazil were the major hub for the global dispersal of the virus. Together, these data provide novel insight into the epidemiological dynamics of this virus and possibly other pandemic viruses.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

