Clearance of HIV infection by selective elimination of host cells capable of producing HIV
- PMID: 32792548
- PMCID: PMC7426846
- DOI: 10.1038/s41467-020-17753-w
Clearance of HIV infection by selective elimination of host cells capable of producing HIV
Erratum in
-
Author Correction: Clearance of HIV infection by selective elimination of host cells capable of producing HIV.Nat Commun. 2020 Dec 2;11(1):6281. doi: 10.1038/s41467-020-20218-9. Nat Commun. 2020. PMID: 33268775 Free PMC article.
Abstract
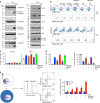
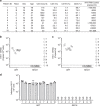
The RNA genome of the human immunodeficiency virus (HIV) is reverse-transcribed into DNA and integrated into the host genome, resulting in latent infections that are difficult to clear. Here we show an approach to eradicate HIV infections by selective elimination of host cells harboring replication-competent HIV (SECH), which includes viral reactivation, induction of cell death, inhibition of autophagy and the blocking of new infections. Viral reactivation triggers cell death specifically in HIV-1-infected T cells, which is promoted by agents that induce apoptosis and inhibit autophagy. SECH treatments can clear HIV-1 in >50% mice reconstituted with a human immune system, as demonstrated by the lack of viral rebound after withdrawal of treatments, and by adoptive transfer of treated lymphocytes into uninfected humanized mice. Moreover, SECH clears HIV-1 in blood samples from HIV-1-infected patients. Our results suggest a strategy to eradicate HIV infections by selectively eliminating host cells capable of producing HIV.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

