Setting a baseline for global urban virome surveillance in sewage
- PMID: 32792677
- PMCID: PMC7426863
- DOI: 10.1038/s41598-020-69869-0
Setting a baseline for global urban virome surveillance in sewage
Erratum in
-
Author Correction: Setting a baseline for global urban virome surveillance in sewage.Sci Rep. 2021 Aug 25;11(1):17446. doi: 10.1038/s41598-021-95934-3. Sci Rep. 2021. PMID: 34433858 Free PMC article. No abstract available.
Abstract
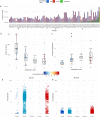
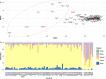
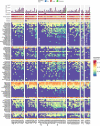
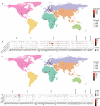
The rapid development of megacities, and their growing connectedness across the world is becoming a distinct driver for emerging disease outbreaks. Early detection of unusual disease emergence and spread should therefore include such cities as part of risk-based surveillance. A catch-all metagenomic sequencing approach of urban sewage could potentially provide an unbiased insight into the dynamics of viral pathogens circulating in a community irrespective of access to care, a potential which already has been proven for the surveillance of poliovirus. Here, we present a detailed characterization of sewage viromes from a snapshot of 81 high density urban areas across the globe, including in-depth assessment of potential biases, as a proof of concept for catch-all viral pathogen surveillance. We show the ability to detect a wide range of viruses and geographical and seasonal differences for specific viral groups. Our findings offer a cross-sectional baseline for further research in viral surveillance from urban sewage samples and place previous studies in a global perspective.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Koopmans M, et al. Familiar barriers still unresolved—a perspective on the Zika virus outbreak research response. Lancet Infect. Dis. 2019;19:e59–e62. - PubMed
-
- Peeling RW, Murtagh M, Olliaro PL. Epidemic preparedness: Why is there a need to accelerate the development of diagnostics? Lancet Infect. Dis. 2019;19:e172–e178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

