Clinical Validation of Two Recombinase-Based Isothermal Amplification Assays (RPA/RAA) for the Rapid Detection of African Swine Fever Virus
- PMID: 32793160
- PMCID: PMC7385304
- DOI: 10.3389/fmicb.2020.01696
Clinical Validation of Two Recombinase-Based Isothermal Amplification Assays (RPA/RAA) for the Rapid Detection of African Swine Fever Virus
Abstract
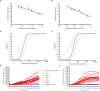
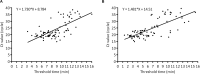
African swine fever (ASF), caused by African swine fever virus (ASFV), is a devastating infectious disease of domestic pigs and wild boars, and has tremendous negative socioeconomic impact on the swine industry and food security worldwide. It is characterized as a notifiable disease by World Organisation for Animal Health (OIE). No effective vaccine or treatment against ASF has so far been available. Early detection and rapid diagnosis are of potential significance to control the spread of ASF. Recombinase-based isothermal amplification assay, recombinase polymerase amplification (RPA) developed by TwistDx (Cambridge, United Kingdom) or recombinase-aided amplification (RAA) by Qitian (Wuxi, China), is becoming a molecular tool for the rapid, specific, and cost-effective identification of multiple pathogens. In this study, we aim to investigate if RPA/RAA can be a potential candidate for on-site, rapid and primary detection of ASFV. A panel of 152 clinical samples previously well-characterized by OIE-recommended qPCR was enrolled in this study, including 20 weak positive (Ct value ≥ 30) samples. This panel was consisted of different types, such as EDTA-blood, spleen, lung, lymph node, kidney, tonsil, liver, brain. We evaluated two recombinase-based isothermal amplification assays, RPA or RAA, by targeting the ASFV B646L gene (p72), and validated the clinical performance in comparison with OIE real-time PCR. Our result showed that the analytical sensitivity of RPA and RAA was as 93.4 and 53.6 copies per reaction, respectively at 95% probability in 16 min, at 39°C. They were universally specific for all 24 genotypes of ASFV and no cross reaction to other pathogens including Classical swine fever virus (CSV), Foot-and-mouth disease virus (FMDV), Pseudorabies virus, Porcine circovirus 2 (PCV2), Porcine Reproductive and respiratory syndrome virus (PPRSV). The results on detection of various kinds of clinical samples indicated an excellent diagnostic agreement between RPA, RAA and OIE real-time PCR method, with the kappa value of 0.960 and 0.973, respectively. Compared to real-time PCR, the specificity of both RPA and RAA was 100% (94.40% ∼ 100%, 95% CI), while the sensitivity was 96.59% (90.36% ∼ 99.29%, 95% CI) and 97.73% (92.03% ∼ 99.72%, 95% CI), respectively. Our data demonstrate that the developed recombinase-based amplification assay (RPA/RAA), promisingly equipped with field-deployable instruments, offers a sensitive and specific platform for the rapid and reliable detection of ASFV, especially in the resource-limited settings for the purpose of screening and surveillance of ASF.
Keywords: African swine fever virus; clinical validation; rapid isothermal amplification; recombinase aided amplification; recombinase polymerase amplification.
Copyright © 2020 Fan, Li, Zhao, Liu, Liu, Wang, Dong, Wang, Chi, Song, Sun, Wang, Ha, Zhao, Bao, Wu and Wang.
Figures

References
-
- Cabada M. M., Malaga J. L., Castellanos-Gonzalez A., Bagwell K. A., Naeger P. A., Rogers H. K., et al. (2017). Recombinase polymerase amplification compared to real-time polymerase chain reaction test for the detection of Fasciola hepatica in Human Stool. Am. J. Trop. Med. Hyg. 96 341–346. 10.4269/ajtmh.16-0601 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

