Iron Supplementation Eliminates Antagonistic Interactions Between Root-Associated Bacteria
- PMID: 32793173
- PMCID: PMC7387576
- DOI: 10.3389/fmicb.2020.01742
Iron Supplementation Eliminates Antagonistic Interactions Between Root-Associated Bacteria
Abstract
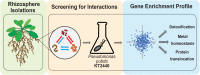
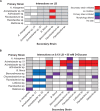
The rhizosphere microbiome (rhizobiome) plays a critical role in plant health and development. However, the processes by which the constituent microbes interact to form and maintain a community are not well understood. To investigate these molecular processes, we examined pairwise interactions between 11 different microbial isolates under select nutrient-rich and nutrient-limited conditions. We observed that when grown with media supplemented with 56 mM glucose, two microbial isolates were able to inhibit the growth of six other microbes. The interaction between microbes persisted even after the antagonistic microbe was removed, upon exposure to spent media. To probe the genetic basis for these antagonistic interactions, we used a barcoded transposon library in a proxy bacterium, Pseudomonas putida, to identify genes which showed enhanced sensitivity to the antagonistic factor(s) secreted by Acinetobacter sp. 02. Iron metabolism-related gene clusters in P. putida were implicated by this systems-level analysis. The supplementation of iron prevented the antagonistic interaction in the original microbial pair, supporting the hypothesis that iron limitation drives antagonistic microbial interactions between rhizobionts. We conclude that rhizobiome community composition is influenced by competition for limiting nutrients, with implications for growth and development of the plant.
Keywords: Acinetobacter; Pseudomonas putida; RB-TnSeq; composition and function; iron depletion; microbe-macroorganism interaction; rhizobiota.
Copyright © 2020 Eng, Herbert, Martinez, Wang, Chen, Brown, Deutschbauer, Bissell, Mortimer and Mukhopadhyay.
Figures




References
-
- Abelson J., Simon M., Guthrie C., Fink G. (1990). Guide to Yeast Genetics and Molecular Biology, Vol194, 1st Edn Amsterdam: Elsevier.

