Geographic and Genomic Distribution of SARS-CoV-2 Mutations
- PMID: 32793182
- PMCID: PMC7387429
- DOI: 10.3389/fmicb.2020.01800
Geographic and Genomic Distribution of SARS-CoV-2 Mutations
Abstract
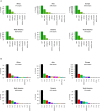
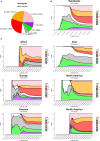
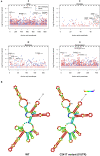
The novel respiratory disease COVID-19 has reached the status of worldwide pandemic and large efforts are currently being undertaken in molecularly characterizing the virus causing it, SARS-CoV-2. The genomic variability of SARS-CoV-2 specimens scattered across the globe can underly geographically specific etiological effects. In the present study, we gather the 48,635 SARS-CoV-2 complete genomes currently available thanks to the collection endeavor of the GISAID consortium and thousands of contributing laboratories. We analyzed and annotated all SARS-CoV-2 mutations compared with the reference Wuhan genome NC_045512.2, observing an average of 7.23 mutations per sample. Our analysis shows the prevalence of single nucleotide transitions as the major mutational type across the world. There exist at least three clades characterized by geographic and genomic specificity. In particular, clade G, prevalent in Europe, carries a D614G mutation in the Spike protein, which is responsible for the initial interaction of the virus with the host human cell. Our analysis may facilitate custom-designed antiviral strategies based on the molecular specificities of SARS-CoV-2 in different patients and geographical locations.
Keywords: COVID-19; SARS-CoV-2; coronavirus; genome evolution; genomics.
Copyright © 2020 Mercatelli and Giorgi.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

