Receptor utilization of angiotensin-converting enzyme 2 (ACE2) indicates a narrower host range of SARS-CoV-2 than that of SARS-CoV
- PMID: 32794346
- PMCID: PMC7436551
- DOI: 10.1111/tbed.13792
Receptor utilization of angiotensin-converting enzyme 2 (ACE2) indicates a narrower host range of SARS-CoV-2 than that of SARS-CoV
Abstract
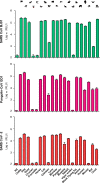
Coronavirus (CoV) pandemics have become a huge threat to the public health worldwide in the recent decades. Typically, severe acute respiratory syndrome CoV (SARS-CoV) caused SARS pandemic in 2003 and SARS-CoV-2 caused the ongoing COVID-19 pandemic. Both viruses are most likely originated from bats. Thus, direct or indirect inter-species transmission from bats to humans is required for the viruses to cause pandemics. Receptor utilization is a key factor determining the host range of viruses which is critical to the inter-species transmission. Angiotensin-converting enzyme 2 (ACE2) is the receptor of both SARS-CoV and SARS-CoV-2, but only ACE2s of certain animals can be utilized by the viruses. Here, we employed pseudovirus cell-entry assay to evaluate the receptor-utilizing capability of ACE2s of 20 animals by the two viruses and found that SARS-CoV-2 utilized less ACE2s than SARS-CoV, indicating a narrower host range of SARS-CoV-2. Especially, SARS-CoV-2 tended not to use murine or non-mammal ACE2s. Meanwhile, pangolin-CoV, another SARS-related coronavirus highly homologous to SARS-CoV-2 in its genome, yet showed similar ACE2 utilization profile with SARS-CoV rather than SARS-CoV-2. Nevertheless, the actual susceptibility of these animals to the coronaviruses should be further verified by in vivo studies. To clarify the mechanism underlying the receptor utilization, we compared the amino acid sequences of the 20 ACE2s and found 5 amino acid residues potentially critical for ACE2 utilization, including the N-terminal 20th and 42nd amino acid residues that might determine the different receptor utilization of SARS-CoV, SARS-CoV-2 and pangolin-CoV. Our studies enhance the understanding of receptor utilization of pandemic coronaviruses, potentially contributing to the virus tracing, intermediate host screening and epidemic prevention for pathogenic coronaviruses.
Keywords: SARS-CoV; SARS-CoV-2; angiotensin-converting enzyme 2 (ACE2); coronavirus; host range; inter-species transmission; receptor utilization.
© 2020 Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interests.
Figures


Similar articles
-
The 442th amino acid residue of the spike protein is critical for the adaptation to bat hosts for SARS-related coronaviruses.Virus Res. 2021 Apr 2;295:198307. doi: 10.1016/j.virusres.2021.198307. Epub 2021 Jan 18. Virus Res. 2021. PMID: 33476695 Free PMC article.
-
Broad and Differential Animal Angiotensin-Converting Enzyme 2 Receptor Usage by SARS-CoV-2.J Virol. 2020 Aug 31;94(18):e00940-20. doi: 10.1128/JVI.00940-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32661139 Free PMC article.
-
Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus.J Virol. 2020 Mar 17;94(7):e00127-20. doi: 10.1128/JVI.00127-20. Print 2020 Mar 17. J Virol. 2020. PMID: 31996437 Free PMC article.
-
Spike Glycoprotein-Mediated Entry of SARS Coronaviruses.Viruses. 2020 Nov 11;12(11):1289. doi: 10.3390/v12111289. Viruses. 2020. PMID: 33187074 Free PMC article. Review.
-
The expression of hACE2 receptor protein and its involvement in SARS-CoV-2 entry, pathogenesis, and its application as potential therapeutic target.Tumour Biol. 2021;43(1):177-196. doi: 10.3233/TUB-200084. Tumour Biol. 2021. PMID: 34420993 Review.
Cited by
-
Variations in Cell Surface ACE2 Levels Alter Direct Binding of SARS-CoV-2 Spike Protein and Viral Infectivity: Implications for Measuring Spike Protein Interactions with Animal ACE2 Orthologs.J Virol. 2022 Sep 14;96(17):e0025622. doi: 10.1128/jvi.00256-22. Epub 2022 Aug 24. J Virol. 2022. PMID: 36000847 Free PMC article.
-
Investigating SARS-CoV-2 Susceptibility in Animal Species: A Scoping Review.Environ Health Insights. 2022 Jun 28;16:11786302221107786. doi: 10.1177/11786302221107786. eCollection 2022. Environ Health Insights. 2022. PMID: 35782319 Free PMC article.
-
The taxonomy, host range and pathogenicity of coronaviruses and other viruses in the Nidovirales order.Anim Dis. 2021;1(1):5. doi: 10.1186/s44149-021-00005-9. Epub 2021 Apr 23. Anim Dis. 2021. PMID: 34778878 Free PMC article. Review.
-
A pair of noncompeting neutralizing human monoclonal antibodies protecting from disease in a SARS-CoV-2 infection model.Eur J Immunol. 2022 May;52(5):770-783. doi: 10.1002/eji.202149374. Epub 2021 Sep 13. Eur J Immunol. 2022. PMID: 34355795 Free PMC article.
-
SARS-CoV-2 surveillance in Norway rats (Rattus norvegicus) from Antwerp sewer system, Belgium.Transbound Emerg Dis. 2022 Sep;69(5):3016-3021. doi: 10.1111/tbed.14219. Epub 2021 Jul 10. Transbound Emerg Dis. 2022. PMID: 34224205 Free PMC article.
References
-
- Ferrario, C. M. , Trask, A. J. , & Jessup, J. A. (2005). Advances in biochemical and functional roles of angiotensin‐converting enzyme 2 and angiotensin‐(1–7) in regulation of cardiovascular function. American Journal of Physiology. Heart and Circulatory Physiology, 289(6), H2281–2290. 10.1152/ajpheart.00618.2005 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 32041001/National Natural Science Foundation of China
- 81902070/National Natural Science Foundation of China
- 2019JJ20004/Provincial Natural Science Foundation of Hunan Province
- 2019JJ50035/Provincial Natural Science Foundation of Hunan Province
- 2020SK3001/Provincial Natural Science Foundation of Hunan Province
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

