High-Resolution Transcriptomic Profiling of the Heart During Chronic Stress Reveals Cellular Drivers of Cardiac Fibrosis and Hypertrophy
- PMID: 32795101
- PMCID: PMC7547893
- DOI: 10.1161/CIRCULATIONAHA.119.045115
High-Resolution Transcriptomic Profiling of the Heart During Chronic Stress Reveals Cellular Drivers of Cardiac Fibrosis and Hypertrophy
Abstract
Background: Cardiac fibrosis is a key antecedent to many types of cardiac dysfunction including heart failure. Physiological factors leading to cardiac fibrosis have been recognized for decades. However, the specific cellular and molecular mediators that drive cardiac fibrosis, and the relative effect of disparate cell populations on cardiac fibrosis, remain unclear.
Methods: We developed a novel cardiac single-cell transcriptomic strategy to characterize the cardiac cellulome, the network of cells that forms the heart. This method was used to profile the cardiac cellular ecosystem in response to 2 weeks of continuous administration of angiotensin II, a profibrotic stimulus that drives pathological cardiac remodeling.
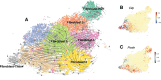
Results: Our analysis provides a comprehensive map of the cardiac cellular landscape uncovering multiple cell populations that contribute to pathological remodeling of the extracellular matrix of the heart. Two phenotypically distinct fibroblast populations, Fibroblast-Cilp and Fibroblast-Thbs4, emerged after induction of tissue stress to promote fibrosis in the absence of smooth muscle actin-expressing myofibroblasts, a key profibrotic cell population. After angiotensin II treatment, Fibroblast-Cilp develops as the most abundant fibroblast subpopulation and the predominant fibrogenic cell type. Mapping intercellular communication networks within the heart, we identified key intercellular trophic relationships and shifts in cellular communication after angiotensin II treatment that promote the development of a profibrotic cellular microenvironment. Furthermore, the cellular responses to angiotensin II and the relative abundance of fibrogenic cells were sexually dimorphic.
Conclusions: These results offer a valuable resource for exploring the cardiac cellular landscape in health and after chronic cardiovascular stress. These data provide insights into the cellular and molecular mechanisms that promote pathological remodeling of the mammalian heart, highlighting early transcriptional changes that precede chronic cardiac fibrosis.
Keywords: fibroblasts; fibrosis; heart failure.
Conflict of interest statement
None.
Figures

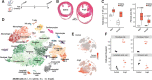




References
-
- Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA, Pinto AR. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 2018;22:600–610. doi: 10.1016/j.celrep.2017.12.072 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

