DPHL: A DIA Pan-human Protein Mass Spectrometry Library for Robust Biomarker Discovery
- PMID: 32795611
- PMCID: PMC7646093
- DOI: 10.1016/j.gpb.2019.11.008
DPHL: A DIA Pan-human Protein Mass Spectrometry Library for Robust Biomarker Discovery
Abstract
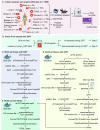
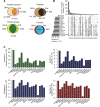
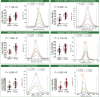
To address the increasing need for detecting and validating protein biomarkers in clinical specimens, mass spectrometry (MS)-based targeted proteomic techniques, including the selected reaction monitoring (SRM), parallel reaction monitoring (PRM), and massively parallel data-independent acquisition (DIA), have been developed. For optimal performance, they require the fragment ion spectra of targeted peptides as prior knowledge. In this report, we describe a MS pipeline and spectral resource to support targeted proteomics studies for human tissue samples. To build the spectral resource, we integrated common open-source MS computational tools to assemble a freely accessible computational workflow based on Docker. We then applied the workflow to generate DPHL, a comprehensive DIA pan-human library, from 1096 data-dependent acquisition (DDA) MS raw files for 16 types of cancer samples. This extensive spectral resource was then applied to a proteomic study of 17 prostate cancer (PCa) patients. Thereafter, PRM validation was applied to a larger study of 57 PCa patients and the differential expression of three proteins in prostate tumor was validated. As a second application, the DPHL spectral resource was applied to a study consisting of plasma samples from 19 diffuse large B cell lymphoma (DLBCL) patients and 18 healthy control subjects. Differentially expressed proteins between DLBCL patients and healthy control subjects were detected by DIA-MS and confirmed by PRM. These data demonstrate that the DPHL supports DIA and PRM MS pipelines for robust protein biomarker discovery. DPHL is freely accessible at https://www.iprox.org/page/project.html?id=IPX0001400000.
Keywords: Data-independent acquisition; Diffuse large B cell lymphoma; Parallel reaction monitoring; Prostate cancer; Spectral library.
Copyright © 2020 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The research group of Tiannan Guo is partly supported by Thermo Fisher Scientific and Pressure Biosciences, which provided access to advanced sample preparation instrumentation. Yue Xuan, Mo Hu, and Yue Zhou were employees of Thermo Fisher Scientific during this project. The remaining authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

