FATTY ACID DESATURASE5 Is Required to Induce Autoimmune Responses in Gigantic Chloroplast Mutants of Arabidopsis
- PMID: 32796124
- PMCID: PMC7534476
- DOI: 10.1105/tpc.20.00016
FATTY ACID DESATURASE5 Is Required to Induce Autoimmune Responses in Gigantic Chloroplast Mutants of Arabidopsis
Abstract
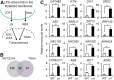
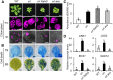
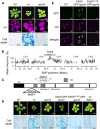
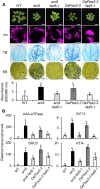
Chloroplasts mediate genetically controlled cell death via chloroplast-to-nucleus retrograde signaling. To decipher the mechanism, we examined chloroplast-linked lesion-mimic mutants of Arabidopsis (Arabidopsis thaliana) deficient in plastid division, thereby developing gigantic chloroplasts (GCs). These GC mutants, including crumpled leaf (crl), constitutively express immune-related genes and show light-dependent localized cell death (LCD), mirroring typical autoimmune responses. Our reverse genetic approach excludes any potential role of immune/stress hormones in triggering LCD. Instead, transcriptome and in silico analyses suggest that reactive electrophile species (RES) generated via oxidation of polyunsaturated fatty acids (PUFAs) or lipid peroxidation-driven signaling may induce LCD. Consistent with these results, the one of the suppressors of crl, dubbed spcrl4, contains a causative mutation in the nuclear gene encoding chloroplast-localized FATTY ACID DESATURASE5 (FAD5) that catalyzes the conversion of palmitic acid (16:0) to palmitoleic acid (16:1). The loss of FAD5 in the crl mutant might attenuate the levels of RES and/or lipid peroxidation due to the reduced levels of palmitic acid-driven PUFAs, which are prime targets of reactive oxygen species. The fact that fad5 also compromises the expression of immune-related genes and the development of LCD in other GC mutants substantiates the presence of an intrinsic retrograde signaling pathway, priming the autoimmune responses in a FAD5-dependent manner.
© 2020 American Society of Plant Biologists. All rights reserved.
Figures



Comment in
-
More Than Just a FAD(5): Unsaturated Fatty Acids in Chloroplasts Elicit Protective Autoimmunity.Plant Cell. 2020 Oct;32(10):3049-3050. doi: 10.1105/tpc.20.00637. Epub 2020 Aug 13. Plant Cell. 2020. PMID: 32796125 Free PMC article. No abstract available.
References
-
- Alméras, E, Stolz, S, Vollenweider, S, Reymond, P, Mène-Saffrané, L., Farmer, E.E(2003). Reactive electrophile species activate defense gene expression in Arabidopsis. Plant J. 34: 205–216. - PubMed
-
- Apel, K, Hirt, H .(2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399. - PubMed
-
- Asano, T, Yoshioka, Y, Kurei, S, Sakamoto, W, Machida, Y; Sodmergen. (2004). A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis. Plant J. 38: 448–459. - PubMed
-
- Baruah, A., Simková, K., Apel, K., Laloi, C.(2009). Arabidopsis mutants reveal multiple singlet oxygen signaling pathways involved in stress response and development. Plant Mol. Biol. 70: 547–563. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

