Development of an NGS-Based Workflow for Improved Monitoring of Circulating Plasmids in Support of Risk Assessment of Antimicrobial Resistance Gene Dissemination
- PMID: 32796589
- PMCID: PMC7460218
- DOI: 10.3390/antibiotics9080503
Development of an NGS-Based Workflow for Improved Monitoring of Circulating Plasmids in Support of Risk Assessment of Antimicrobial Resistance Gene Dissemination
Abstract
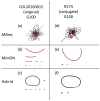
Antimicrobial resistance (AMR) is one of the most prominent public health threats. AMR genes localized on plasmids can be easily transferred between bacterial isolates by horizontal gene transfer, thereby contributing to the spread of AMR. Next-generation sequencing (NGS) technologies are ideal for the detection of AMR genes; however, reliable reconstruction of plasmids is still a challenge due to large repetitive regions. This study proposes a workflow to reconstruct plasmids with NGS data in view of AMR gene localization, i.e., chromosomal or on a plasmid. Whole-genome and plasmid DNA extraction methods were compared, as were assemblies consisting of short reads (Illumina MiSeq), long reads (Oxford Nanopore Technologies) and a combination of both (hybrid). Furthermore, the added value of conjugation of a plasmid to a known host was evaluated. As a case study, an isolate harboring a large, low-copy mcr-1-carrying plasmid (>200 kb) was used. Hybrid assemblies of NGS data obtained from whole-genome DNA extractions of the original isolates resulted in the most complete reconstruction of plasmids. The optimal workflow was successfully applied to multidrug-resistant Salmonella Kentucky isolates, where the transfer of an ESBL-gene-containing fragment from a plasmid to the chromosome was detected. This study highlights a strategy including wet and dry lab parameters that allows accurate plasmid reconstruction, which will contribute to an improved monitoring of circulating plasmids and the assessment of their risk of transfer.
Keywords: DNA extraction; Flongle; MiSeq; MinION; antimicrobial resistance; conjugation; hybrid assembly; mobile elements; next-generation sequencing; plasmids; surveillance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- WHO . Antimicrobial Resistance: Global Report on Surveillance 2014. WHO Press; Geneva, Switzerland: 2014.
-
- Trieu-Cuot P., Carlier C., Martin P., Courvalin P. Plasmid transfer by conjugation from Escherichia coli to Gram-positive bacteria. FEMS Microbiol. Lett. 1987;48:289–294. doi: 10.1111/j.1574-6968.1987.tb02558.x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

