Footprint of the host restriction factors APOBEC3 on the genome of human viruses
- PMID: 32797103
- PMCID: PMC7449416
- DOI: 10.1371/journal.ppat.1008718
Footprint of the host restriction factors APOBEC3 on the genome of human viruses
Abstract
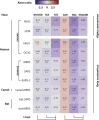
APOBEC3 enzymes are innate immune effectors that introduce mutations into viral genomes. These enzymes are cytidine deaminases which transform cytosine into uracil. They preferentially mutate cytidine preceded by thymidine making the 5'TC motif their favored target. Viruses have evolved different strategies to evade APOBEC3 restriction. Certain viruses actively encode viral proteins antagonizing the APOBEC3s, others passively face the APOBEC3 selection pressure thanks to a depleted genome for APOBEC3-targeted motifs. Hence, the APOBEC3s left on the genome of certain viruses an evolutionary footprint. The aim of our study is the identification of these viruses having a genome shaped by the APOBEC3s. We analyzed the genome of 33,400 human viruses for the depletion of APOBEC3-favored motifs. We demonstrate that the APOBEC3 selection pressure impacts at least 22% of all currently annotated human viral species. The papillomaviridae and polyomaviridae are the most intensively footprinted families; evidencing a selection pressure acting genome-wide and on both strands. Members of the parvoviridae family are differentially targeted in term of both magnitude and localization of the footprint. Interestingly, a massive APOBEC3 footprint is present on both strands of the B19 erythroparvovirus; making this viral genome one of the most cleaned sequences for APOBEC3-favored motifs. We also identified the endemic coronaviridae as significantly footprinted. Interestingly, no such footprint has been detected on the zoonotic MERS-CoV, SARS-CoV-1 and SARS-CoV-2 coronaviruses. In addition to viruses that are footprinted genome-wide, certain viruses are footprinted only on very short sections of their genome. That is the case for the gamma-herpesviridae and adenoviridae where the footprint is localized on the lytic origins of replication. A mild footprint can also be detected on the negative strand of the reverse transcribing HIV-1, HIV-2, HTLV-1 and HBV viruses. Together, our data illustrate the extent of the APOBEC3 selection pressure on the human viruses and identify new putatively APOBEC3-targeted viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures










References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

