FANCJ compensates for RAP80 deficiency and suppresses genomic instability induced by interstrand cross-links
- PMID: 32797166
- PMCID: PMC7498338
- DOI: 10.1093/nar/gkaa660
FANCJ compensates for RAP80 deficiency and suppresses genomic instability induced by interstrand cross-links
Abstract
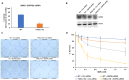
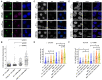
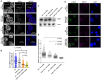
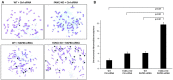
FANCJ, a DNA helicase and interacting partner of the tumor suppressor BRCA1, is crucial for the repair of DNA interstrand crosslinks (ICL), a highly toxic lesion that leads to chromosomal instability and perturbs normal transcription. In diploid cells, FANCJ is believed to operate in homologous recombination (HR) repair of DNA double-strand breaks (DSB); however, its precise role and molecular mechanism is poorly understood. Moreover, compensatory mechanisms of ICL resistance when FANCJ is deficient have not been explored. In this work, we conducted a siRNA screen to identify genes of the DNA damage response/DNA repair regime that when acutely depleted sensitize FANCJ CRISPR knockout cells to a low concentration of the DNA cross-linking agent mitomycin C (MMC). One of the top hits from the screen was RAP80, a protein that recruits repair machinery to broken DNA ends and regulates DNA end-processing. Concomitant loss of FANCJ and RAP80 not only accentuates DNA damage levels in human cells but also adversely affects the cell cycle checkpoint, resulting in profound chromosomal instability. Genetic complementation experiments demonstrated that both FANCJ's catalytic activity and interaction with BRCA1 are important for ICL resistance when RAP80 is deficient. The elevated RPA and RAD51 foci in cells co-deficient of FANCJ and RAP80 exposed to MMC are attributed to single-stranded DNA created by Mre11 and CtIP nucleases. Altogether, our cell-based findings together with biochemical studies suggest a critical function of FANCJ to suppress incompletely processed and toxic joint DNA molecules during repair of ICL-induced DNA damage.
Published by Oxford University Press on behalf of Nucleic Acids Research 2020.
Figures



References
-
- Kohn K.W. Beyond DNA cross-linking: history and prospects of DNA-targeted cancer treatment–fifteenth Bruce F. Cain Memorial Award Lecture. Cancer Res. 1996; 56:5533–5546. - PubMed
-
- Voulgaridou G.P., Anestopoulos I., Franco R., Panayiotidis M.I., Pappa A.. DNA damage induced by endogenous aldehydes: current state of knowledge. Mutat. Res. 2011; 711:13–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

