GPCR Genes as Activators of Surface Colonization Pathways in a Model Marine Diatom
- PMID: 32798972
- PMCID: PMC7452957
- DOI: 10.1016/j.isci.2020.101424
GPCR Genes as Activators of Surface Colonization Pathways in a Model Marine Diatom
Abstract
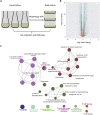
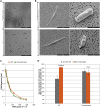
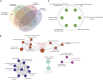
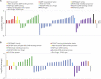
Surface colonization allows diatoms, a dominant group of phytoplankton in oceans, to adapt to harsh marine environments while mediating biofoulings to human-made underwater facilities. The regulatory pathways underlying diatom surface colonization, which involves morphotype switching in some species, remain mostly unknown. Here, we describe the identification of 61 signaling genes, including G-protein-coupled receptors (GPCRs) and protein kinases, which are differentially regulated during surface colonization in the model diatom species, Phaeodactylum tricornutum. We show that the transformation of P. tricornutum with constructs expressing individual GPCR genes induces cells to adopt the surface colonization morphology. P. tricornutum cells transformed to express GPCR1A display 30% more resistance to UV light exposure than their non-biofouling wild-type counterparts, consistent with increased silicification of cell walls associated with the oval biofouling morphotype. Our results provide a mechanistic definition of morphological shifts during surface colonization and identify candidate target proteins for the screening of eco-friendly, anti-biofouling molecules.
Keywords: Genetics; Microbiology.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing financial interests.
Figures


Similar articles
-
Protocol to generate and characterize biofouling transformants of a model marine diatom.STAR Protoc. 2021 Aug 5;2(3):100716. doi: 10.1016/j.xpro.2021.100716. eCollection 2021 Sep 17. STAR Protoc. 2021. PMID: 34401782 Free PMC article.
-
Characterization of marine diatom-infecting virus promoters in the model diatom Phaeodactylum tricornutum.Sci Rep. 2015 Dec 22;5:18708. doi: 10.1038/srep18708. Sci Rep. 2015. PMID: 26692124 Free PMC article.
-
How marine diatoms cope with metal challenge: Insights from the morphotype-dependent metal tolerance in Phaeodactylum tricornutum.Ecotoxicol Environ Saf. 2021 Jan 15;208:111715. doi: 10.1016/j.ecoenv.2020.111715. Epub 2020 Nov 27. Ecotoxicol Environ Saf. 2021. PMID: 33396046
-
Modelling metabolism of the diatom Phaeodactylum tricornutum.Biochem Soc Trans. 2015 Dec;43(6):1182-6. doi: 10.1042/BST20150152. Biochem Soc Trans. 2015. PMID: 26614658 Review.
-
Diatom elemental and morphological changes in response to iron limitation: a brief review with potential paleoceanographic applications.Geobiology. 2009 Sep;7(4):419-31. doi: 10.1111/j.1472-4669.2009.00207.x. Epub 2009 Jul 29. Geobiology. 2009. PMID: 19659798 Review.
Cited by
-
Protocol to generate and characterize biofouling transformants of a model marine diatom.STAR Protoc. 2021 Aug 5;2(3):100716. doi: 10.1016/j.xpro.2021.100716. eCollection 2021 Sep 17. STAR Protoc. 2021. PMID: 34401782 Free PMC article.
-
In vivo thrombin activity in the diatom Phaeodactylum tricornutum: biotechnological insights.Appl Microbiol Biotechnol. 2024 Oct 8;108(1):481. doi: 10.1007/s00253-024-13322-z. Appl Microbiol Biotechnol. 2024. PMID: 39377797 Free PMC article.
-
Genome analysis of Parmales, the sister group of diatoms, reveals the evolutionary specialization of diatoms from phago-mixotrophs to photoautotrophs.Commun Biol. 2023 Jul 7;6(1):697. doi: 10.1038/s42003-023-05002-x. Commun Biol. 2023. PMID: 37420035 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

