Synchronised infection identifies early rate-limiting steps in the hepatitis B virus life cycle
- PMID: 32799415
- PMCID: PMC7611726
- DOI: 10.1111/cmi.13250
Synchronised infection identifies early rate-limiting steps in the hepatitis B virus life cycle
Abstract
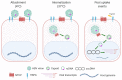
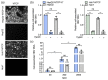
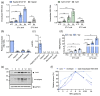
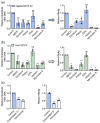
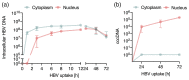
Hepatitis B virus (HBV) is an enveloped DNA virus that contains a partially double-stranded relaxed circular (rc) DNA. Upon infection, rcDNA is delivered to the nucleus where it is repaired to covalently closed circular (ccc) DNA that serves as the transcription template for all viral RNAs. Our understanding of HBV particle entry dynamics and host pathways regulating intracellular virus trafficking and cccDNA formation is limited. The discovery of sodium taurocholate co-transporting peptide (NTCP) as the primary receptor allows studies on these early steps in viral life cycle. We employed a synchronised infection protocol to quantify HBV entry kinetics. HBV attachment to cells at 4°C is independent of NTCP, however, subsequent particle uptake is NTCP-dependent and reaches saturation at 12 h post-infection. HBV uptake is clathrin- and dynamin dependent with actin and tubulin playing a role in the first 6 h of infection. Cellular fractionation studies demonstrate HBV DNA in the nucleus within 6 h of infection and cccDNA was first detected at 24 h post-infection. Our studies show the majority (83%) of cell bound particles enter HepG2-NTCP cells, however, only a minority (<1%) of intracellular rcDNA was converted to cccDNA, highlighting this as a rate-limiting in establishing infection in vitro. This knowledge highlights the deficiencies in our in vitro cell culture systems and will inform the design and evaluation of physiologically relevant models that support efficient HBV replication.
Keywords: hepatitis B; kinetics; virus internalisation.
© 2020 The Authors. Cellular Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no potential conflict of interest.
Figures
References
-
- Arudchandran R, Brown MJ, Song JS, Wank SA, Haleem-Smith H, Rivera J. Polyethylene glycol-mediated infection of non-permissive mammalian cells with Semliki forest virus: Application to signal transduction studies. Journal of Immunological Methods. 1999;222(1-2):197–208. doi: 10.1016/s0022-1759(98)00161-6. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

